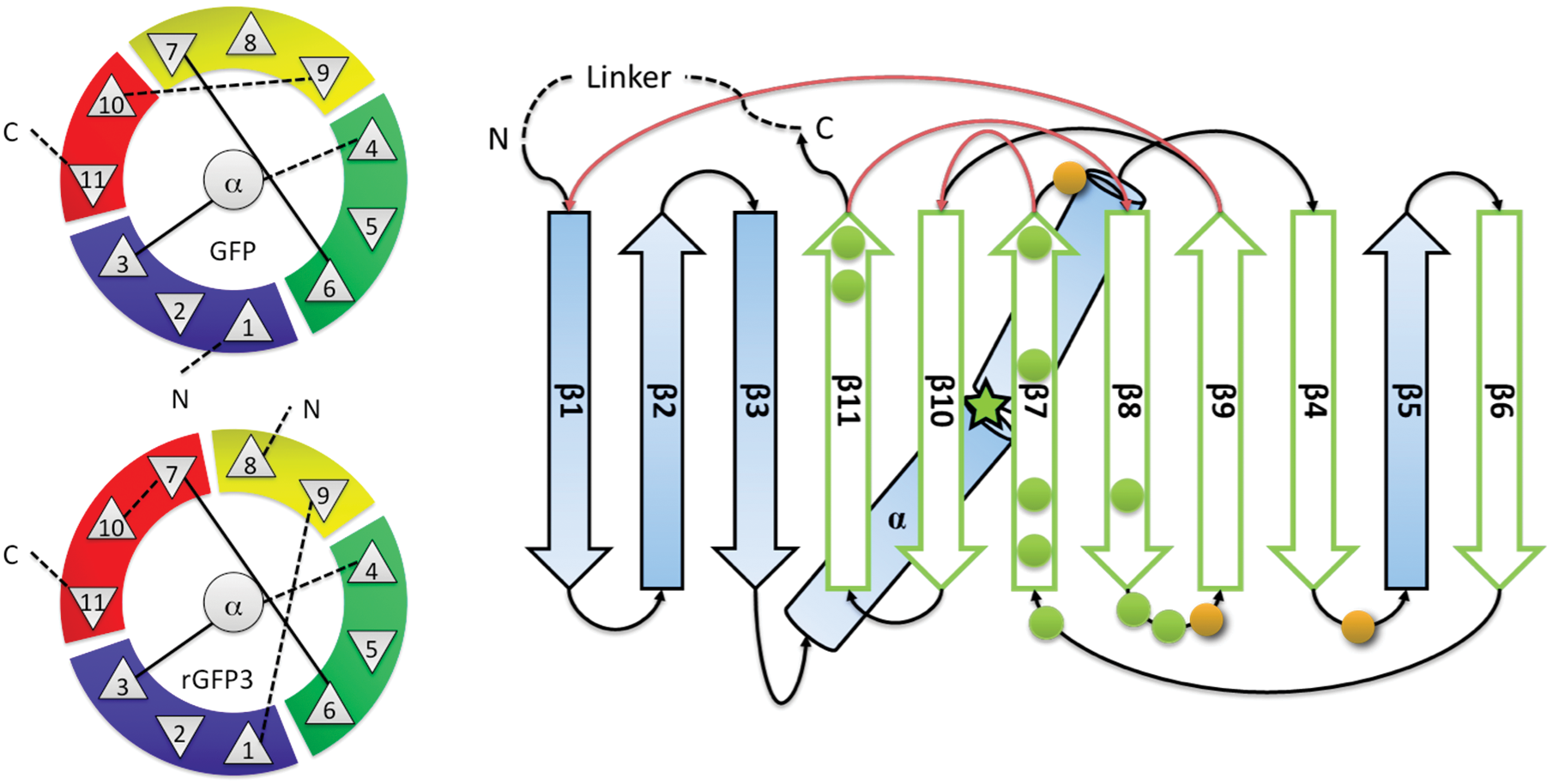
Amino acid sequences of GFP, GFPuv, mutated GFPuv (K79A/R80A-SDM I) and... | Download Scientific Diagram

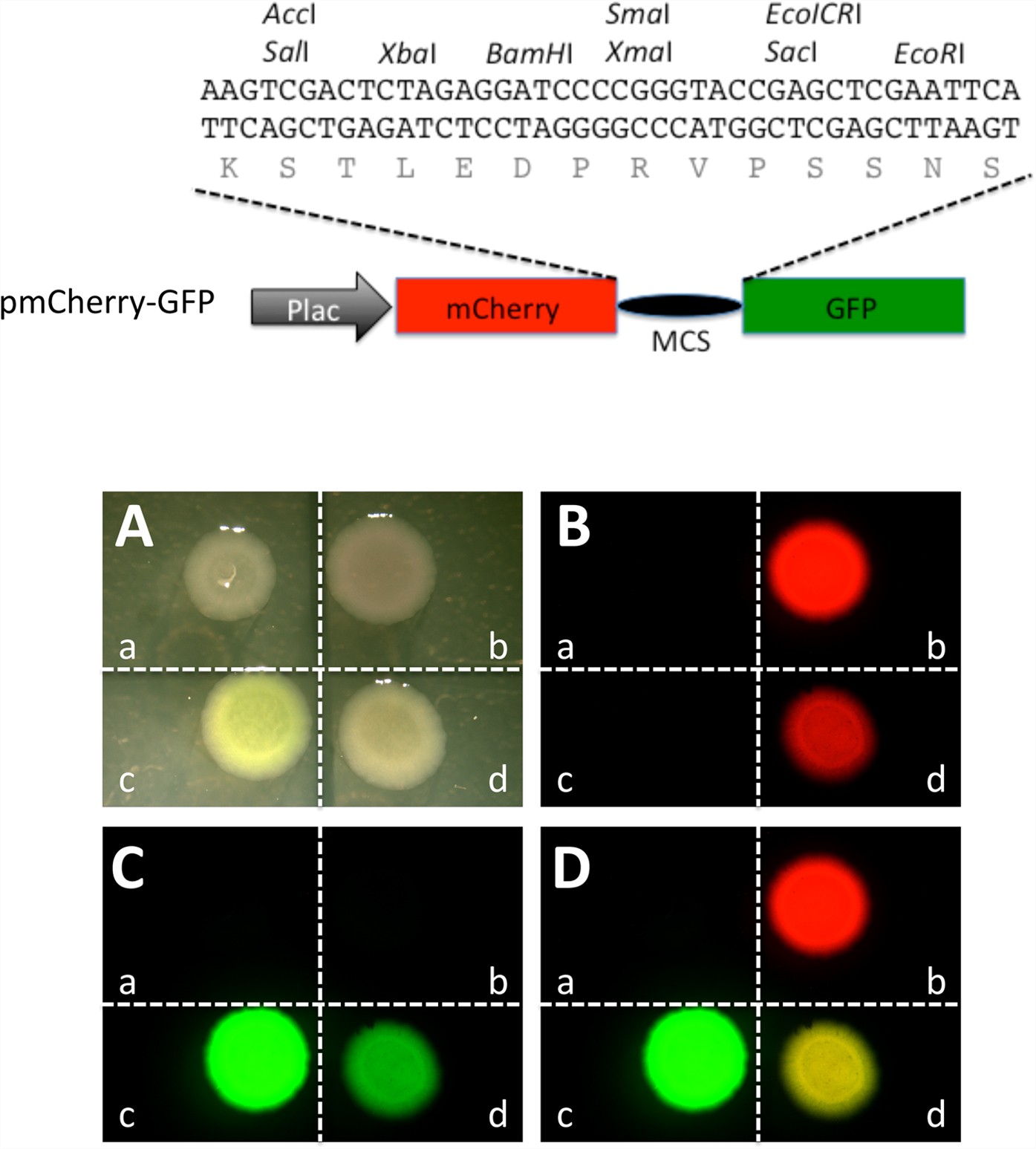
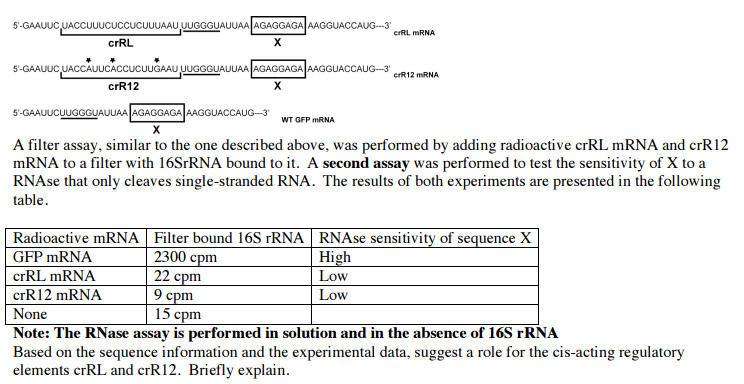
Modified gfp sequences. (A) Schematic diagrams showing gene expression... | Download Scientific Diagram
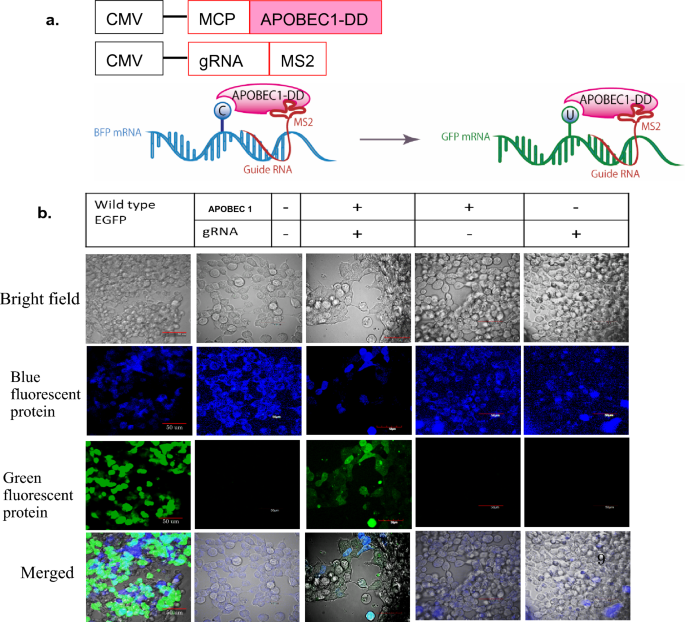
RNA editing of BFP, a point mutant of GFP, using artificial APOBEC1 deaminase to restore the genetic code | Scientific Reports

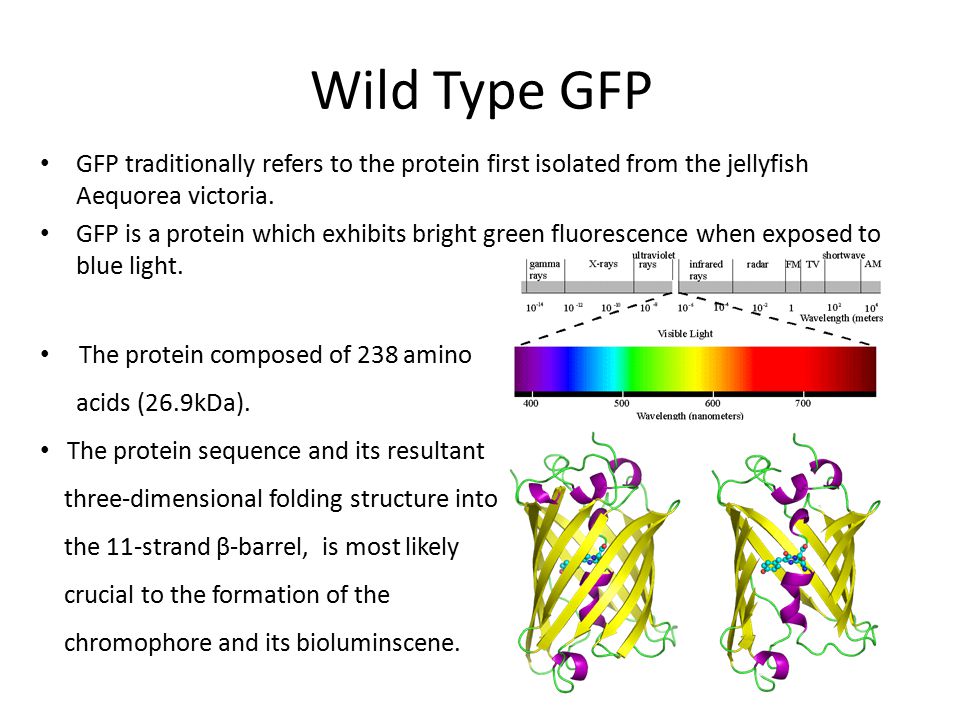
Random GFP∷cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency | PNAS

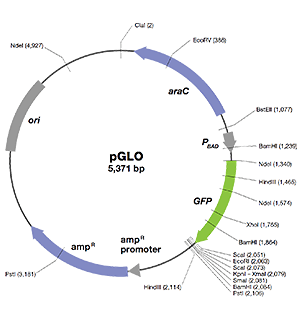
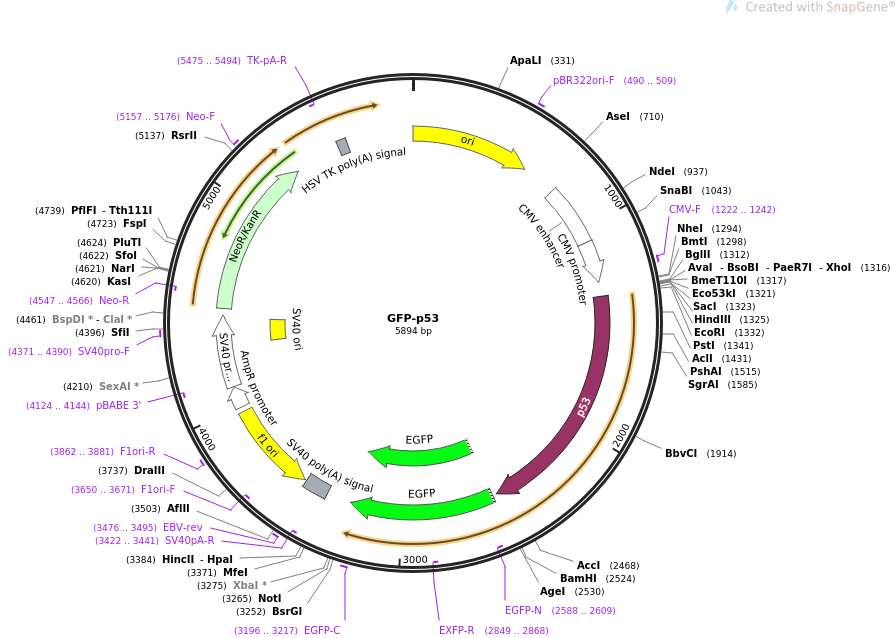
Wild-type psiRNA-hH1GFPzeo G2 vector map showing / restriction sites (*), the α-peptide coding sequence located downstream of the H1 promoter, and the GFP coding sequence located downstream of hEF1-HTLV promoter

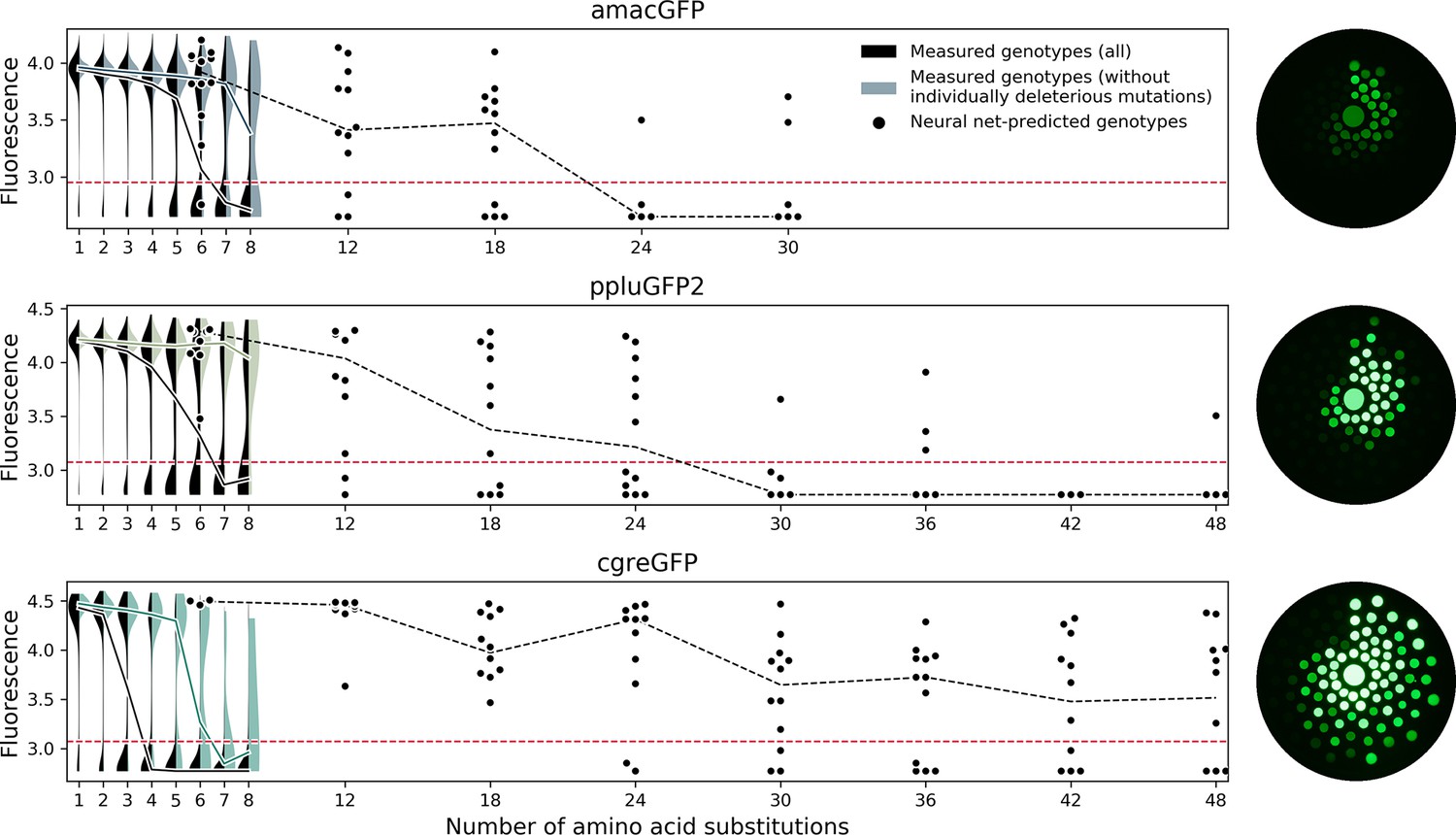
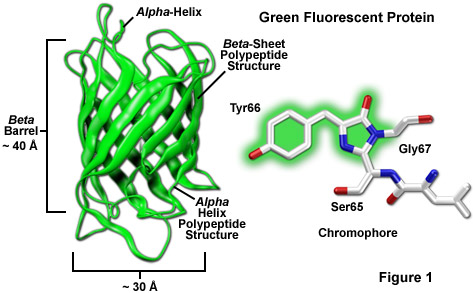
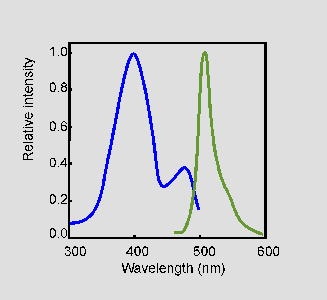
Light-Driven Decarboxylation of Wild-Type Green Fluorescent Protein | Journal of the American Chemical Society

Sequences and genealogy.(a) Sequence alignment of mRFP variants with... | Download Scientific Diagram

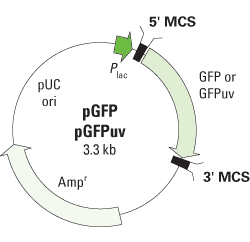
Wild-type and mutant GFP sequences and ssODN design. (A) The pGFPmut... | Download Scientific Diagram

IJMS | Free Full-Text | Studying the Function of Phytoplasma Effector Proteins Using a Chemical-Inducible Expression System in Transgenic Plants

Light-Driven Decarboxylation of Wild-Type Green Fluorescent Protein | Journal of the American Chemical Society