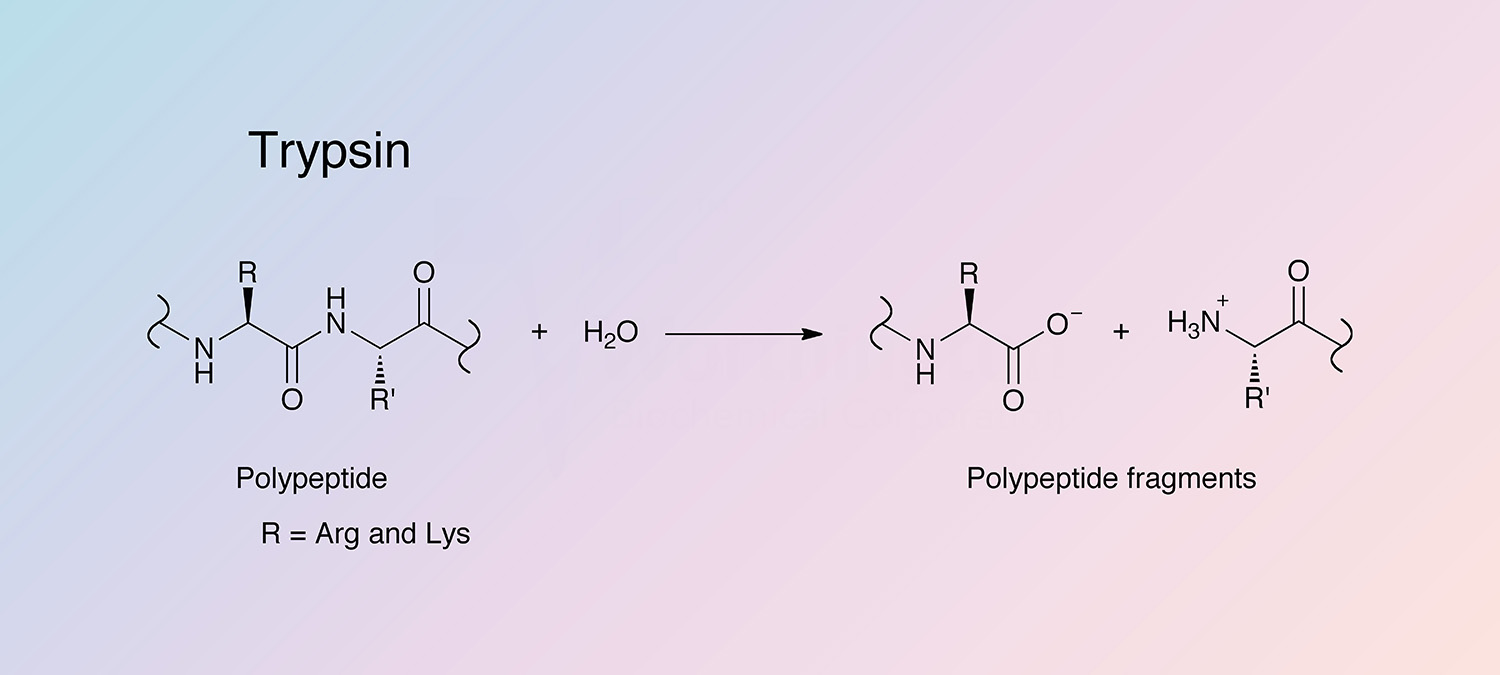
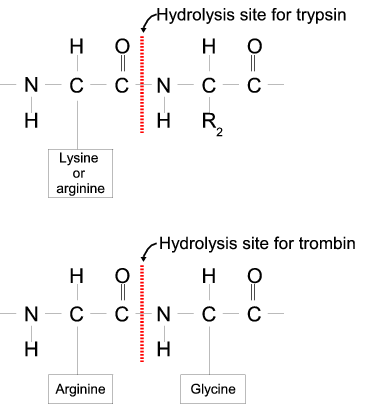
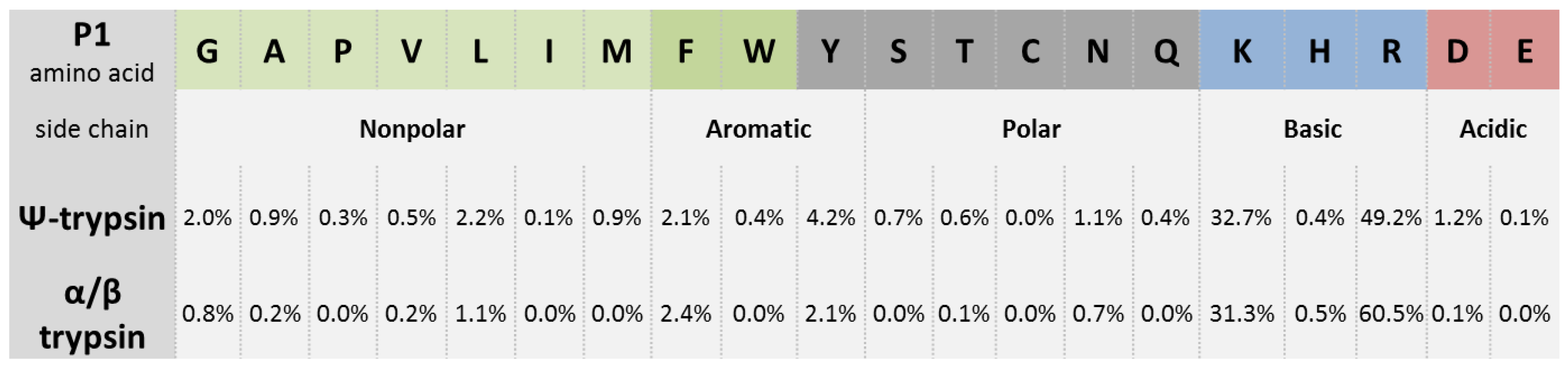
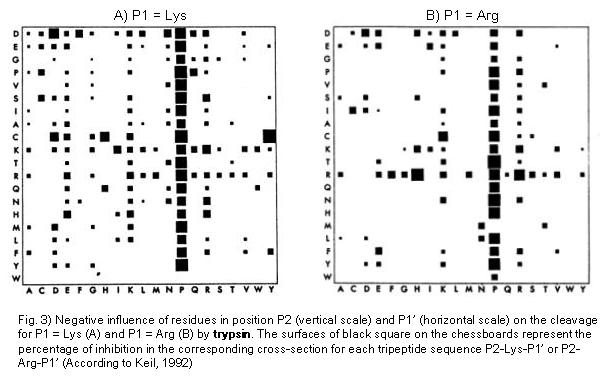
Peptide mapping and amino acid sequence of MalS. Trypsin cleavage sites... | Download Scientific Diagram

PAR2 N-terminus with major cleavage sites identified. N-terminus of... | Download Scientific Diagram
![Sequence of HRPc [1]. The trypsin cleavage sites are indicated by bold... | Download Scientific Diagram Sequence of HRPc [1]. The trypsin cleavage sites are indicated by bold... | Download Scientific Diagram](https://www.researchgate.net/publication/13465969/figure/fig1/AS:602031375003649@1520546873782/Sequence-of-HRPc-1-The-trypsin-cleavage-sites-are-indicated-by-bold-letters-The.png)
Sequence of HRPc [1]. The trypsin cleavage sites are indicated by bold... | Download Scientific Diagram

Nipah virus fusion protein: Influence of cleavage site mutations on the cleavability by cathepsin L, trypsin and furin | Semantic Scholar

Schematic representation of the major trypsin cleavage sites and one... | Download Scientific Diagram

The role of furin cleavage site in SARS-CoV-2 spike protein-mediated membrane fusion in the presence or absence of trypsin | Signal Transduction and Targeted Therapy

Substrate cleavage specificity of legumain, GluC, and trypsin. IceLogos... | Download Scientific Diagram
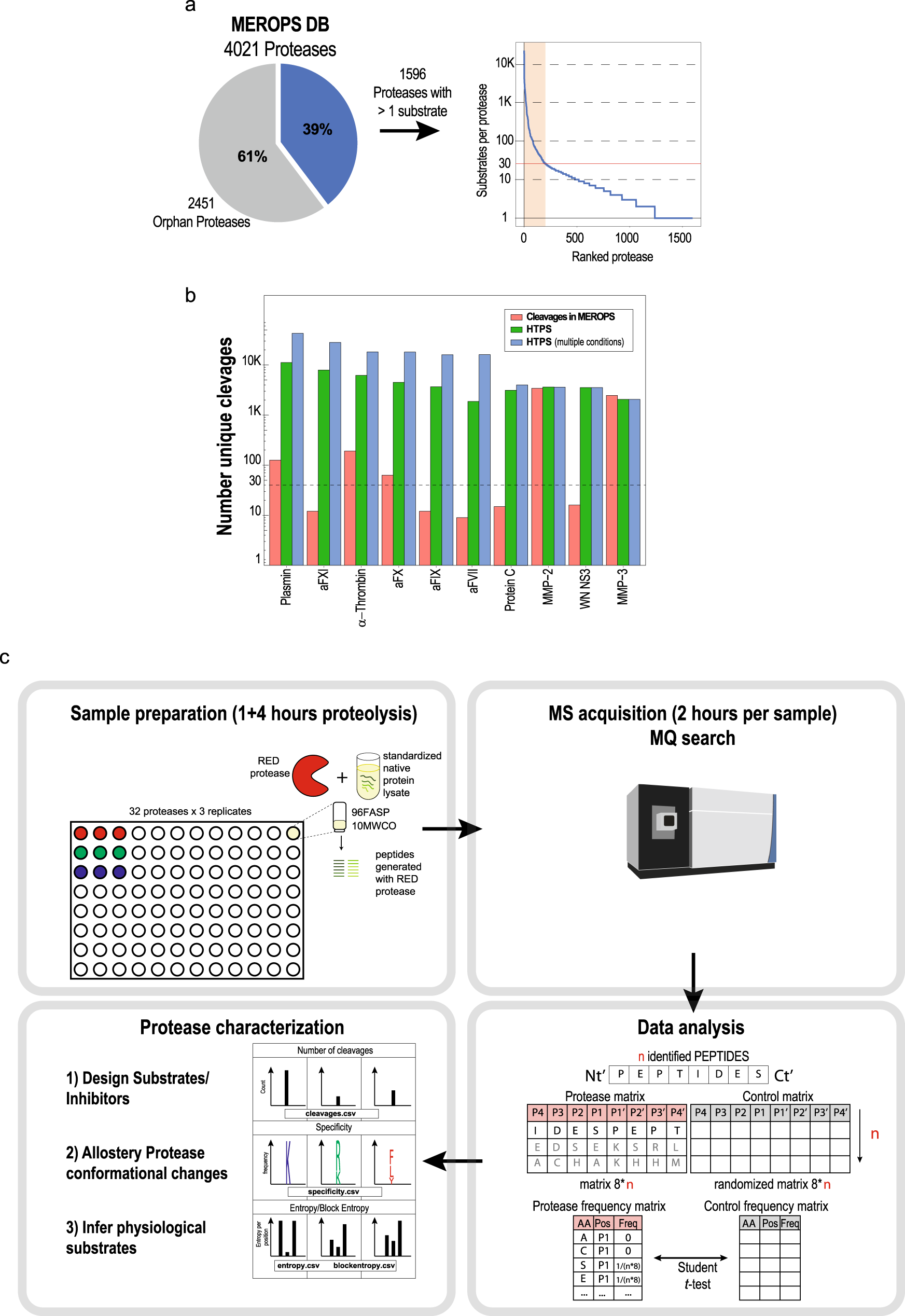
Mapping specificity, cleavage entropy, allosteric changes and substrates of blood proteases in a high-throughput screen | Nature Communications
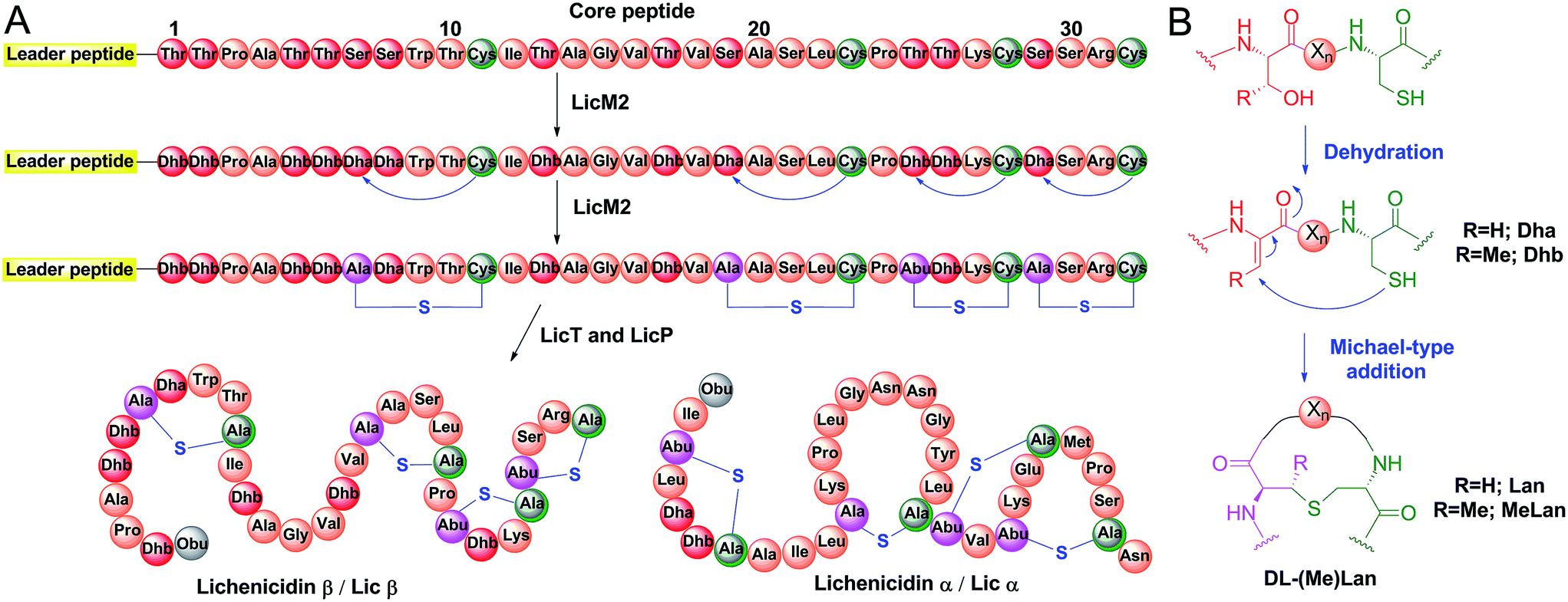
Applications of the class II lanthipeptide protease LicP for sequence-specific, traceless peptide bond cleavage - Chemical Science (RSC Publishing) DOI:10.1039/C5SC02329G

Cleaved and Missed Sites for Trypsin, Lys-C, and Lys-N Can Be Predicted with High Confidence on the Basis of Sequence Context | Journal of Proteome Research
Frontiers | Identification of Cleavage Sites Recognized by the 3C-Like Cysteine Protease within the Two Polyproteins of Strawberry Mottle Virus

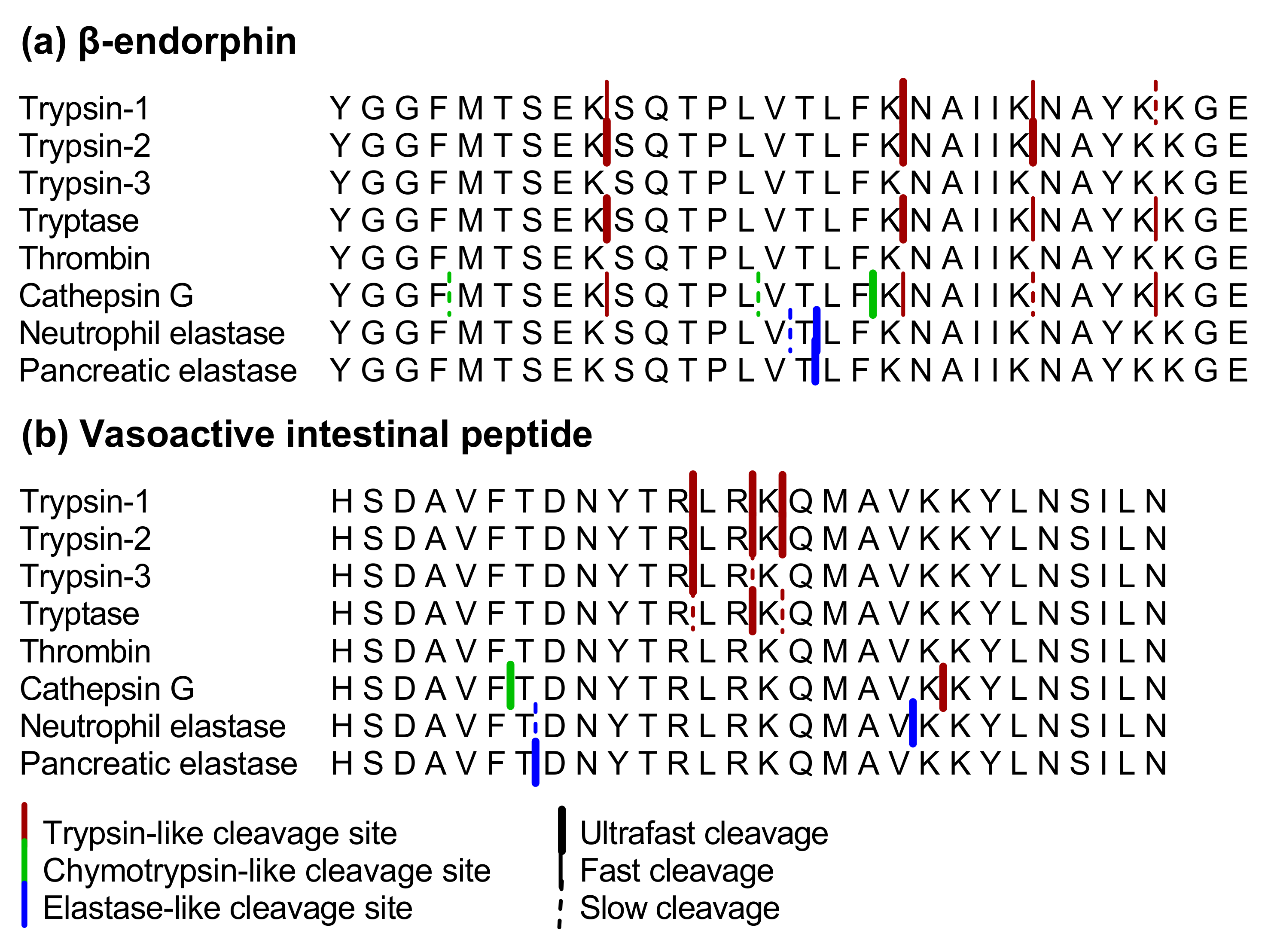
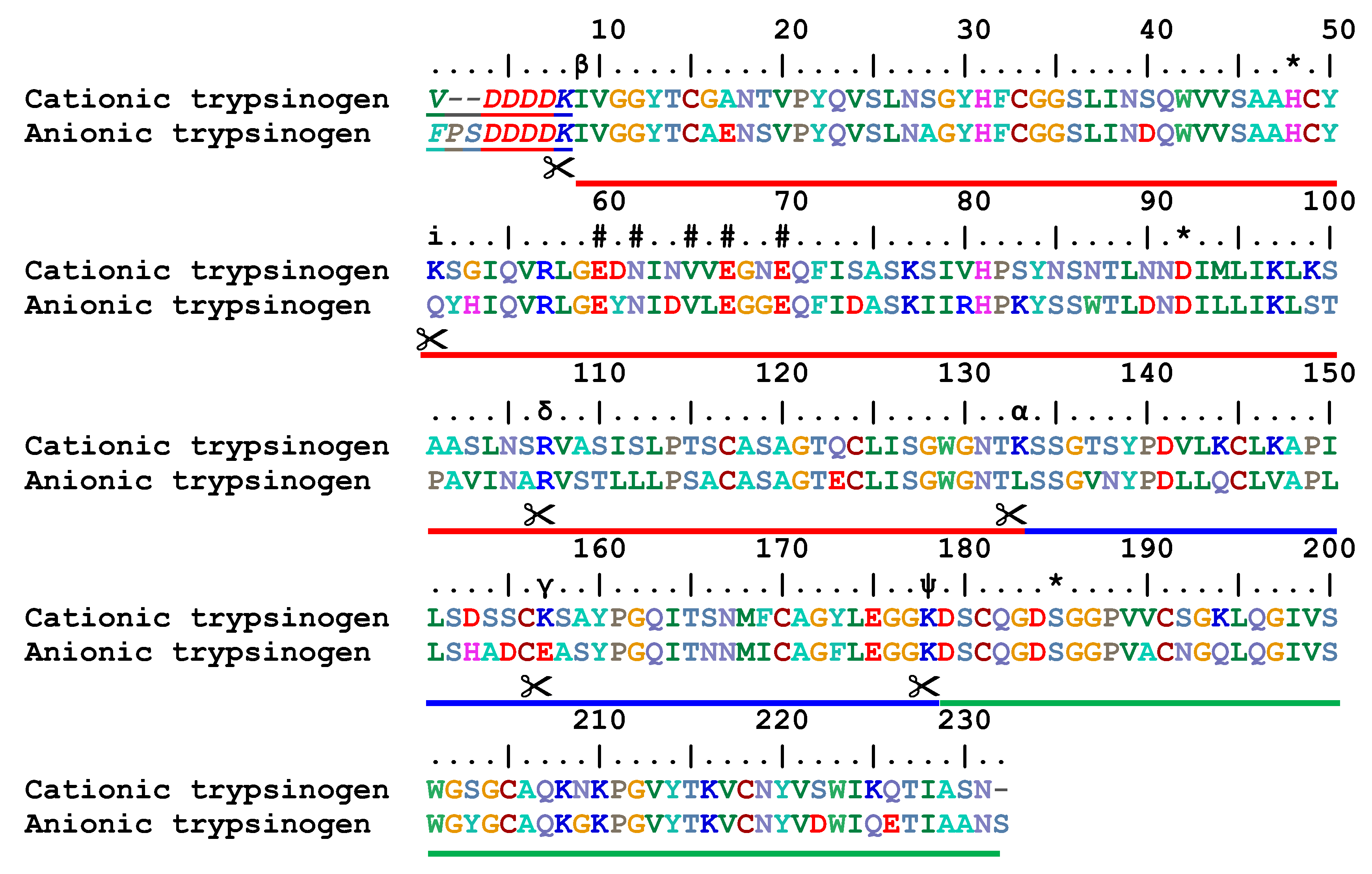
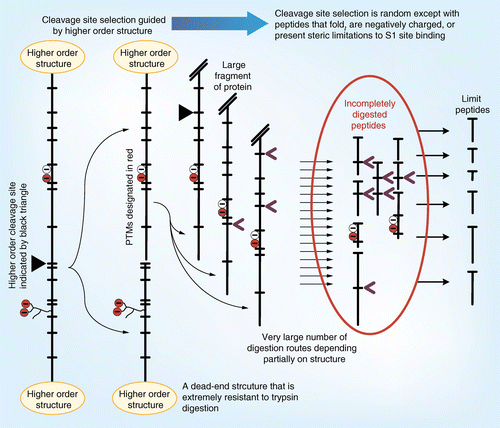
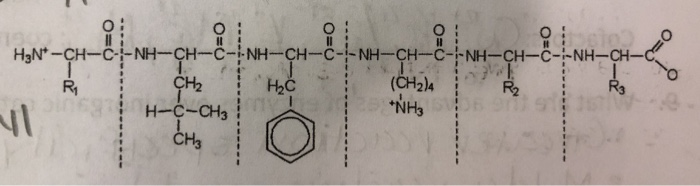
Marked difference in efficiency of the digestive enzymes pepsin, trypsin, chymotrypsin, and pancreatic elastase to cleave tightly folded proteins

Protease cleavage site fingerprinting by label‐free in‐gel degradomics reveals pH‐dependent specificity switch of legumain | The EMBO Journal