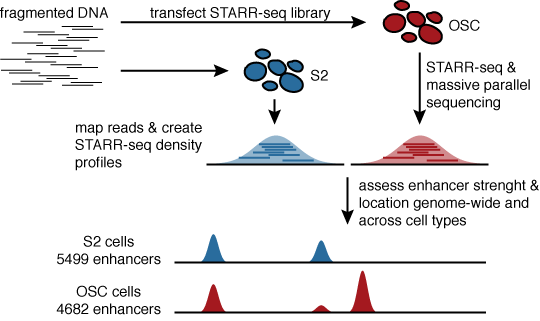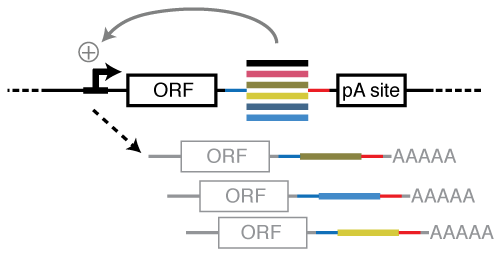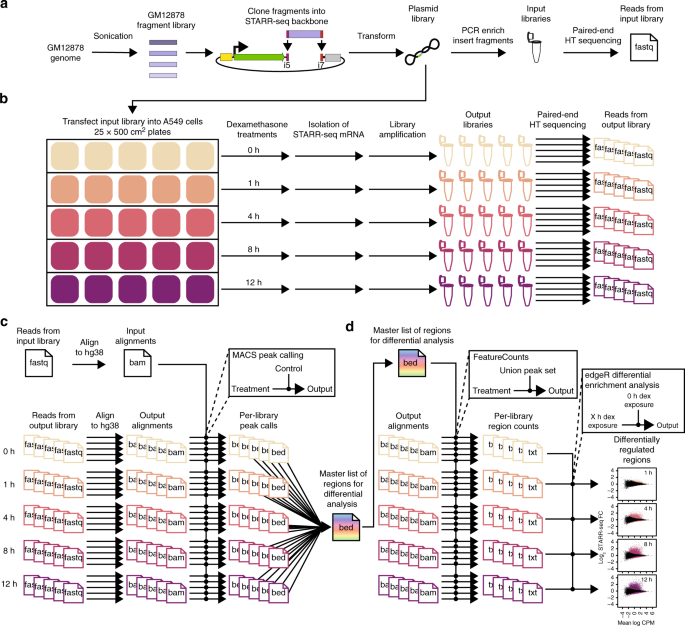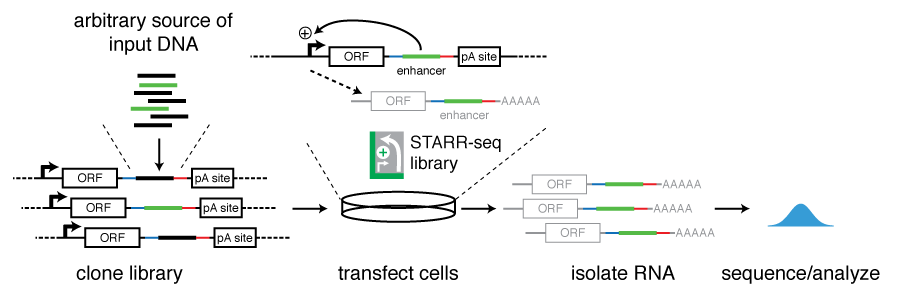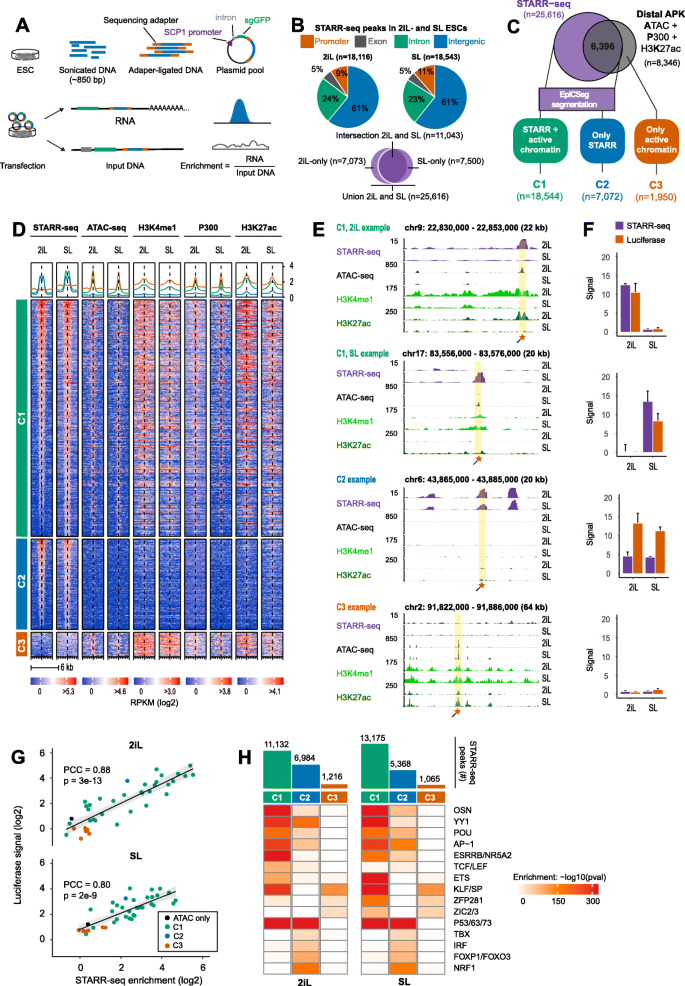
STARR-seq identifies active, chromatin-masked, and dormant enhancers in pluripotent mouse embryonic stem cells | Genome Biology | Full Text
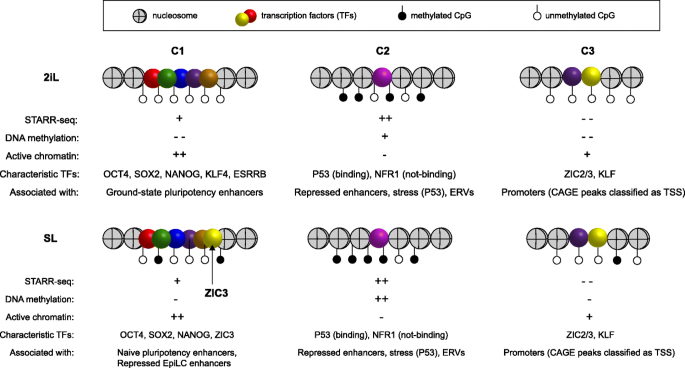
STARR-seq identifies active, chromatin-masked, and dormant enhancers in pluripotent mouse embryonic stem cells | Genome Biology | Full Text
Recent advances in high-throughput approaches to dissect enhancer function[version 1; peer review: 3 approved]

High-throughput and quantitative assessment of enhancer activity in mammals by CapStarr-seq | Nature Communications

High-resolution genome-wide functional dissection of transcriptional regulatory regions and nucleotides in human | Nature Communications
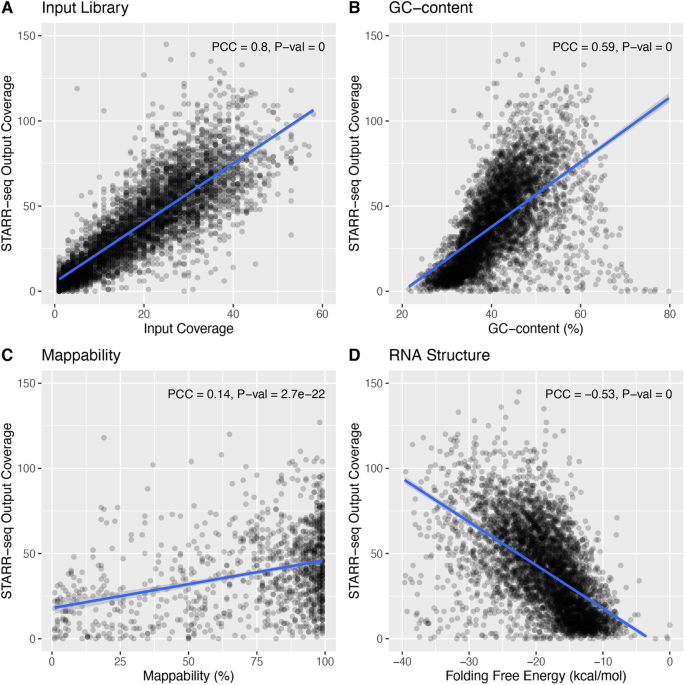
STARRPeaker: uniform processing and accurate identification of STARR-seq active regions | Genome Biology | Full Text
Synthetic STARR-seq reveals how DNA shape and sequence modulate transcriptional output and noise | PLOS Genetics

STARR‐seq and UMI‐STARR‐seq: Assessing Enhancer Activities for Genome‐Wide‐, High‐, and Low‐Complexity Candidate Libraries - Neumayr - 2019 - Current Protocols in Molecular Biology - Wiley Online Library

STARR-seq output coverage is fit against simulated coverage using three... | Download Scientific Diagram
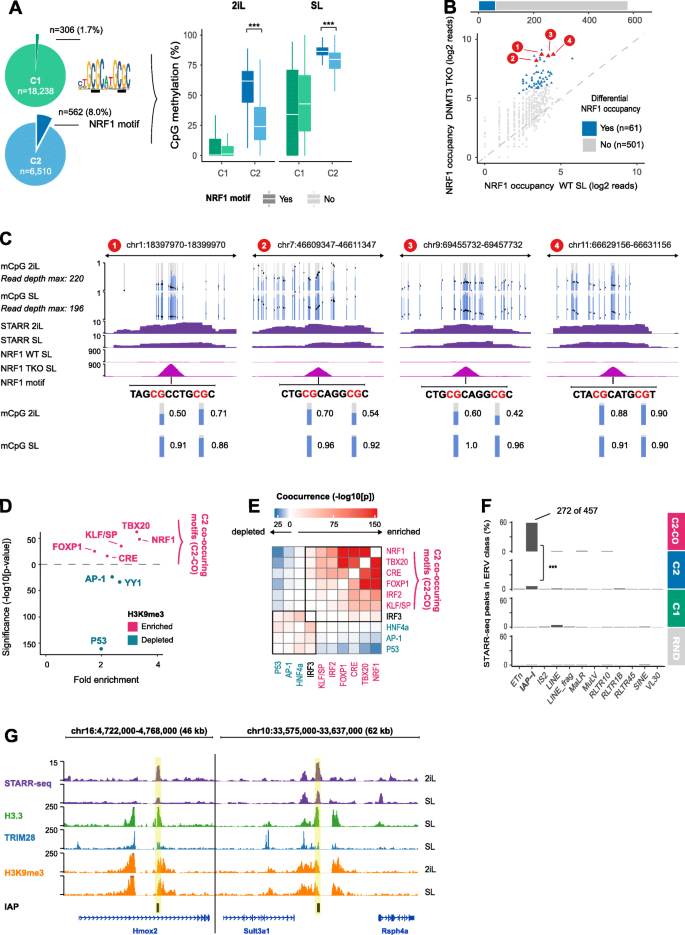
STARR-seq identifies active, chromatin-masked, and dormant enhancers in pluripotent mouse embryonic stem cells | Genome Biology | Full Text

STARR‐seq and UMI‐STARR‐seq: Assessing Enhancer Activities for Genome‐Wide‐, High‐, and Low‐Complexity Candidate Libraries - Neumayr - 2019 - Current Protocols in Molecular Biology - Wiley Online Library

Identification of Functional Variant Enhancers Associated With Atrial Fibrillation | Circulation Research

Identifying transcription factor-bound activators and silencers in the chromatin accessible human genome using ATAC-STARR-seq | bioRxiv

Roadmap for a successful STARR-seq assay: Schematic illustrates five... | Download Scientific Diagram

ExP STARR-seq a, Schematic of the ExP STARR-seq method used to measure... | Download Scientific Diagram