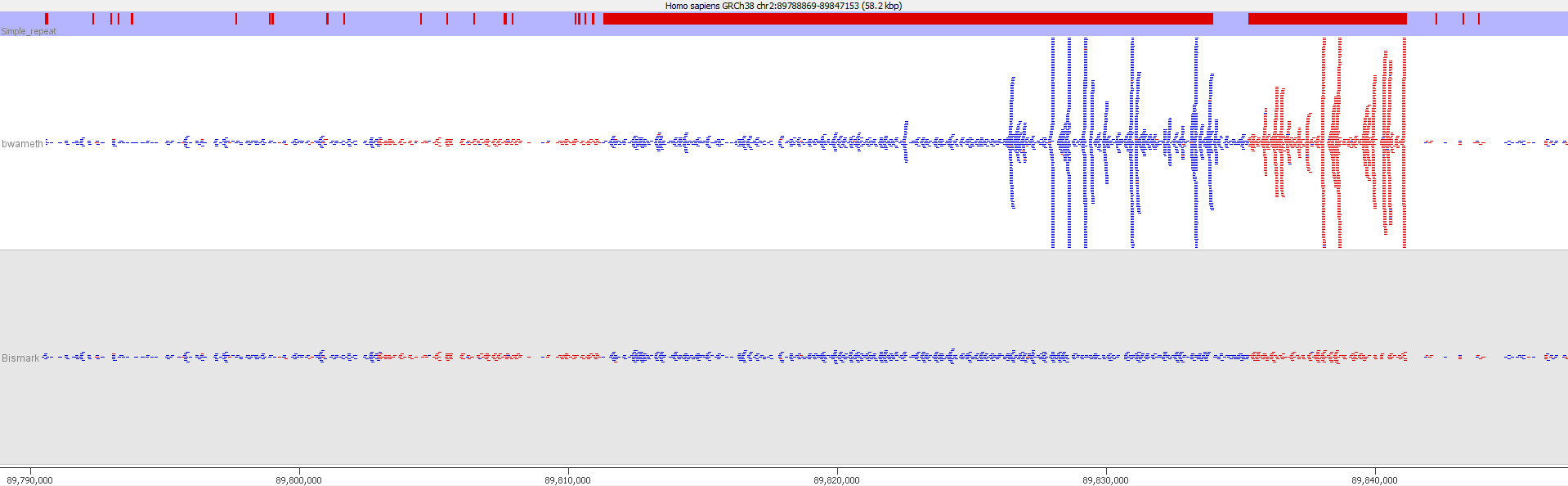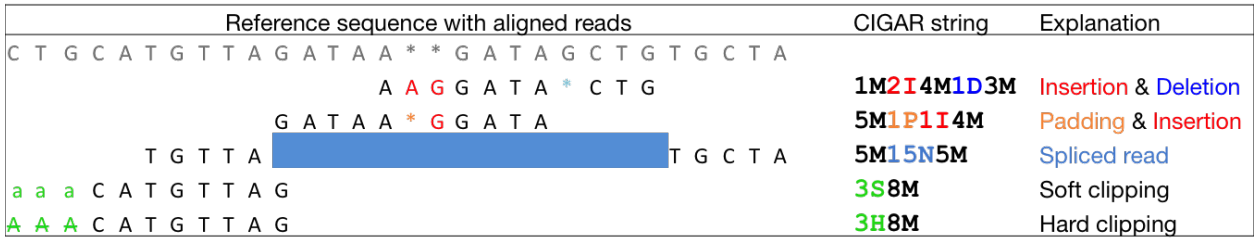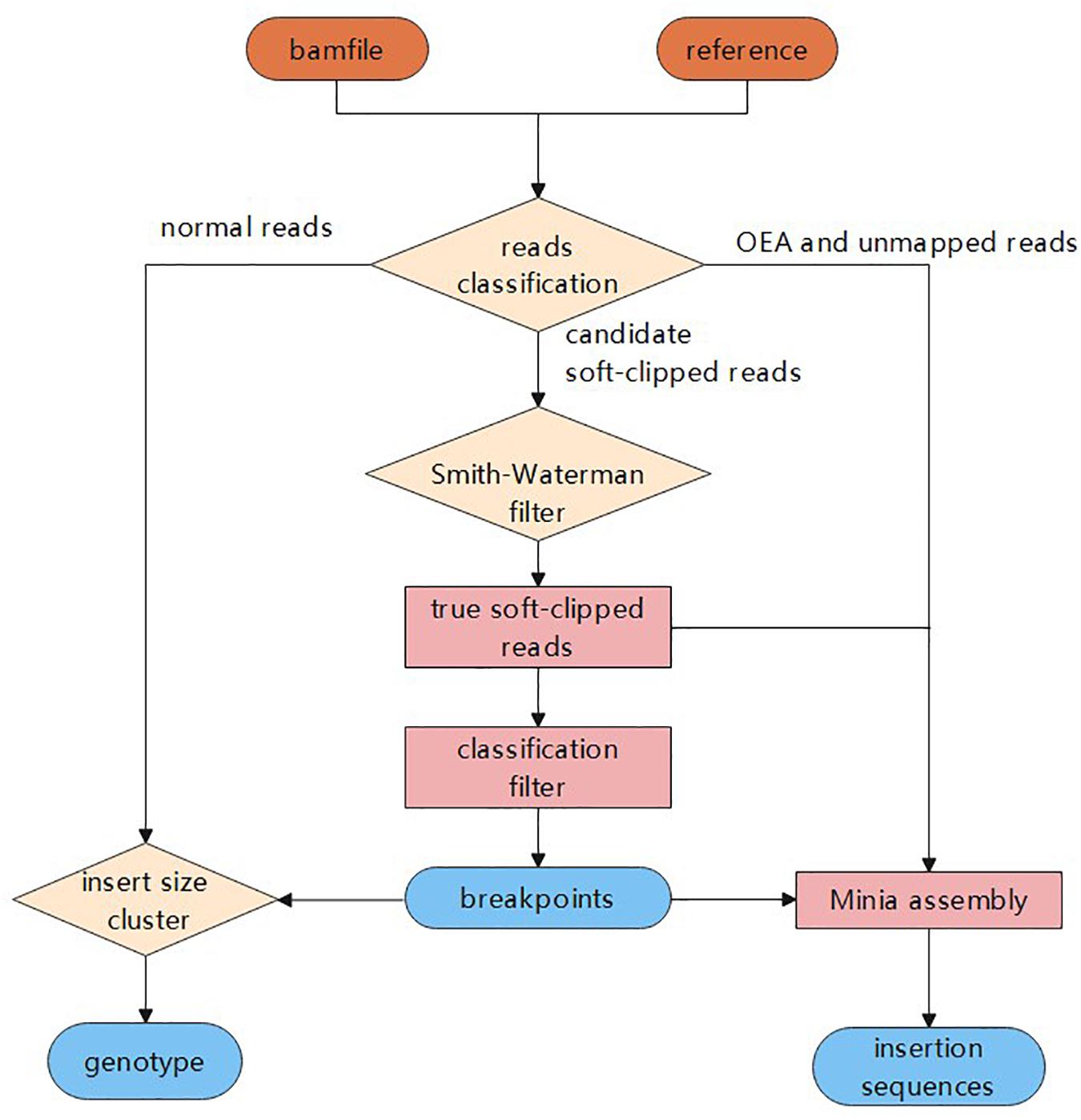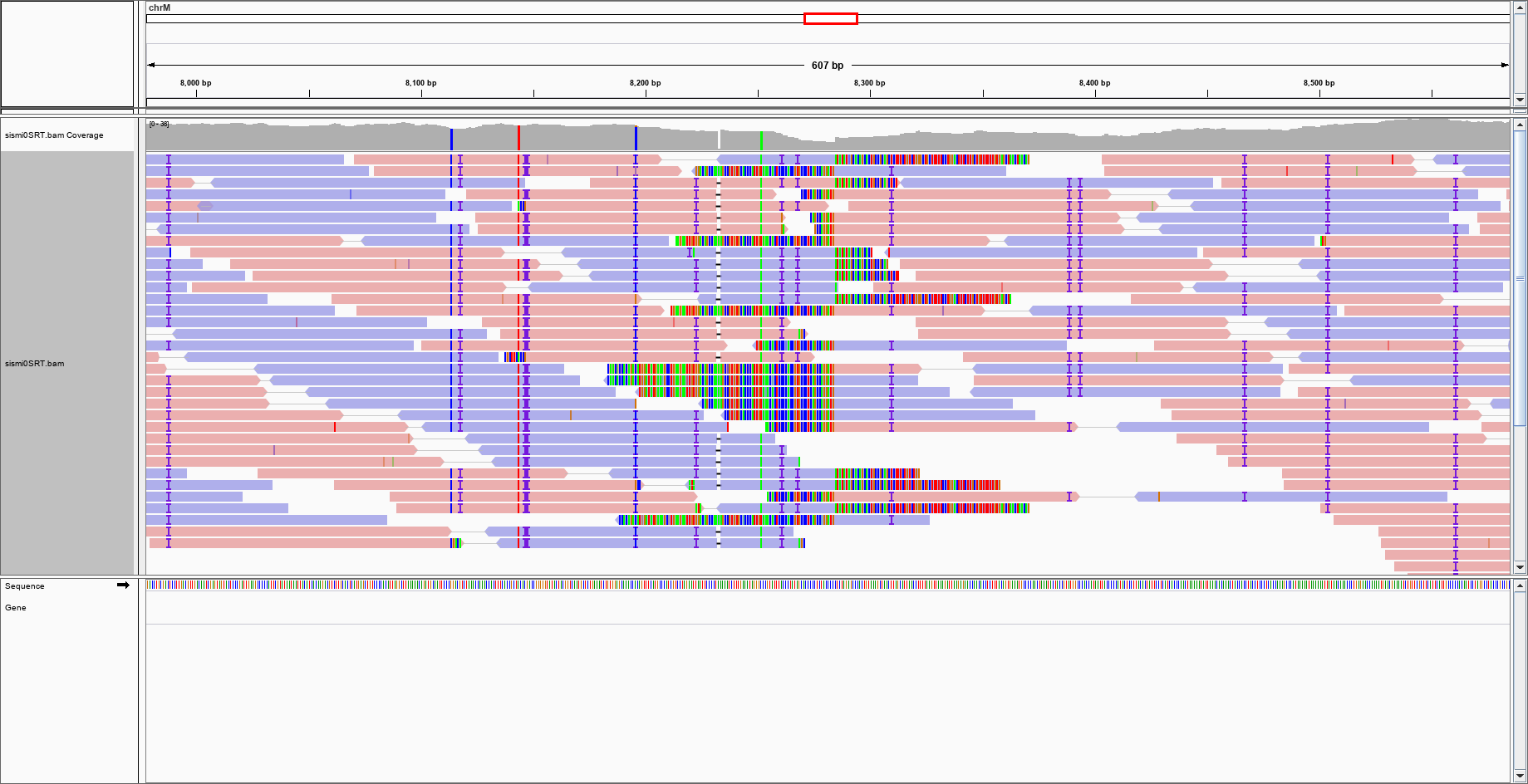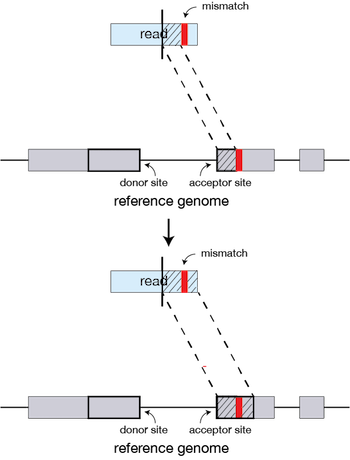
QC Fail Sequencing » Soft-clipping of reads may add potentially unwanted alignments to repetitive regions

Soft-clipped reads vs. improperly aligned reads at a structural variant... | Download Scientific Diagram
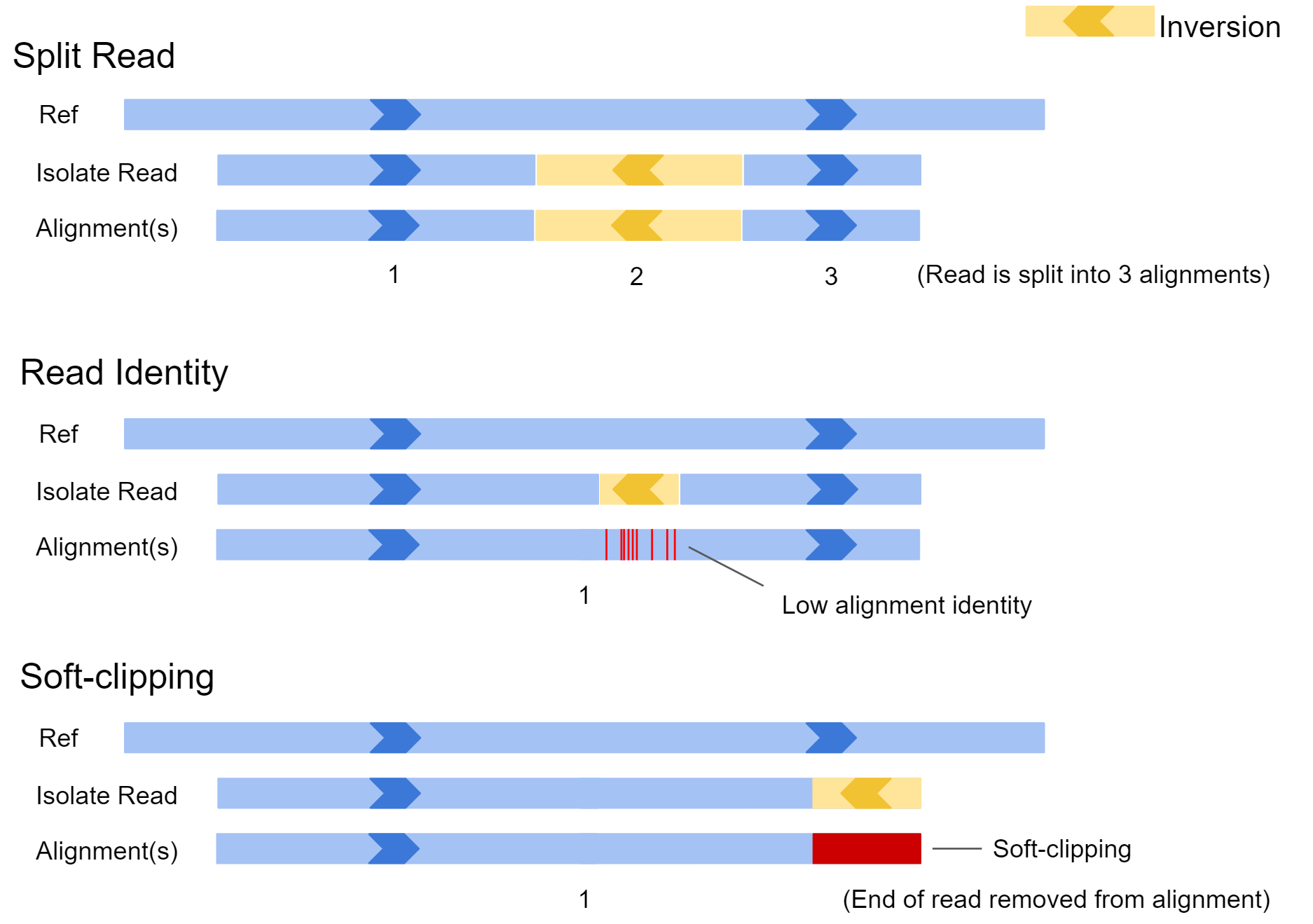
QC Fail Sequencing » Soft-clipping of reads may add potentially unwanted alignments to repetitive regions

eKLIPse: a sensitive tool for the detection and quantification of mitochondrial DNA deletions from next-generation sequencing data - ScienceDirect

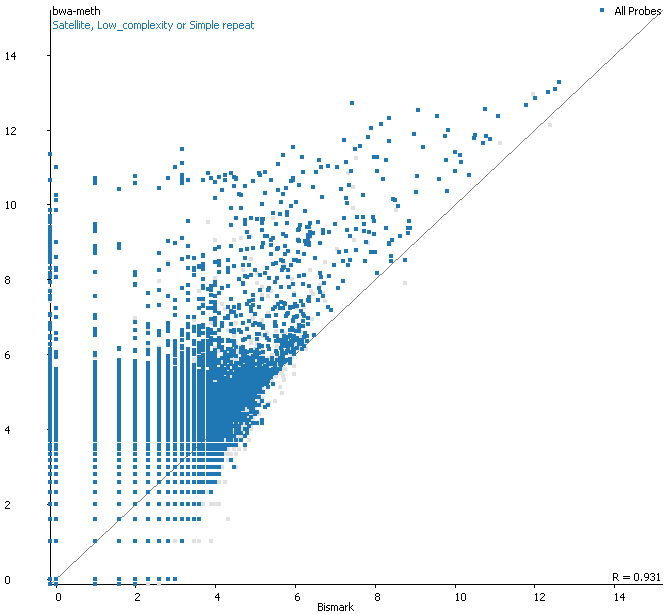
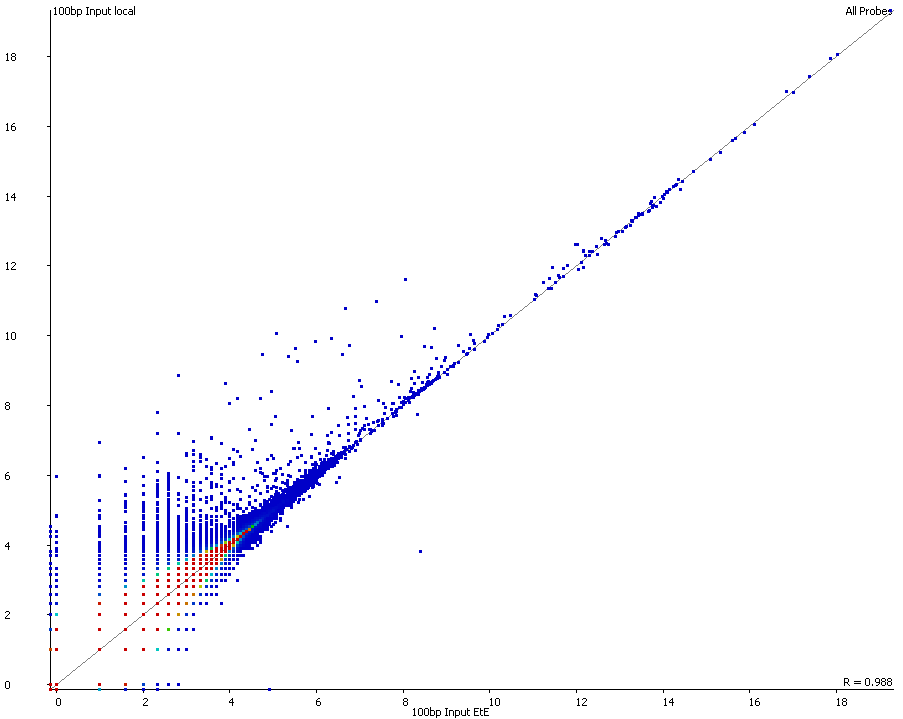
Strange differences in detected variants called by mutect2 --dont-use-soft- clipped-bases true vs false – GATK

Soft-clipped reads vs. improperly aligned reads at a structural variant... | Download Scientific Diagram
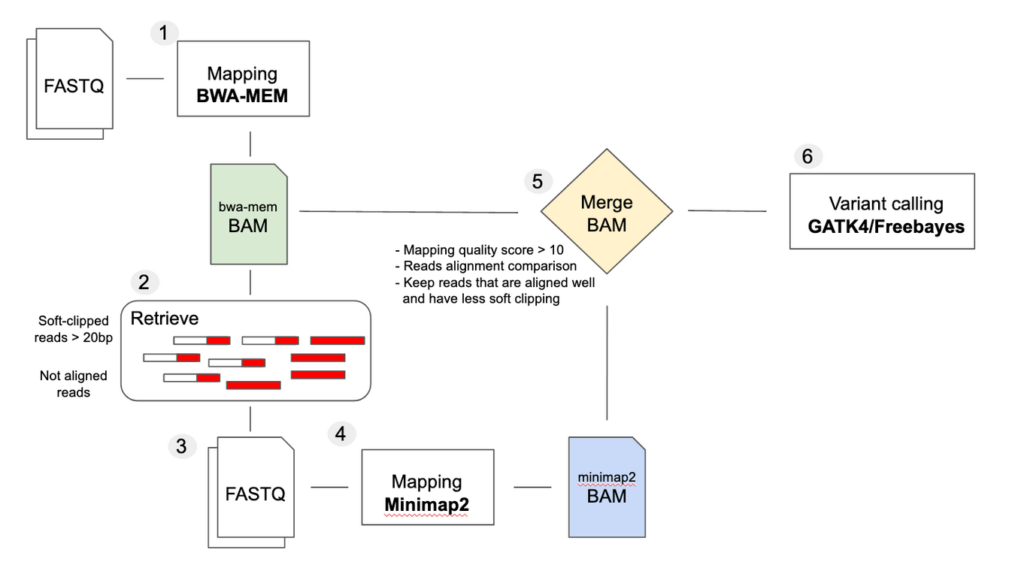
Challenges and importance of mid-sized deletion identification for genomic medicine - SeqOne Genomics

Common sources of 5′end soft clipping and mismatches in 3′ end seq. (a)... | Download Scientific Diagram

Soft-clipped reads vs. improperly aligned reads at a structural variant... | Download Scientific Diagram
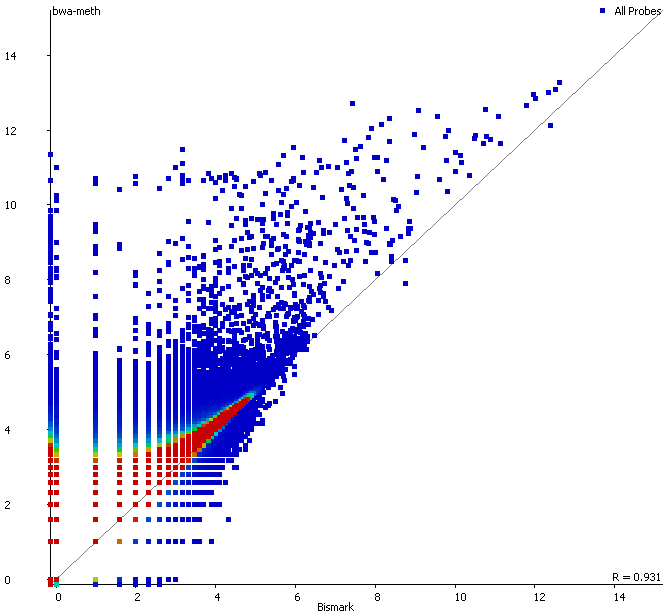
QC Fail Sequencing » Soft-clipping of reads may add potentially unwanted alignments to repetitive regions
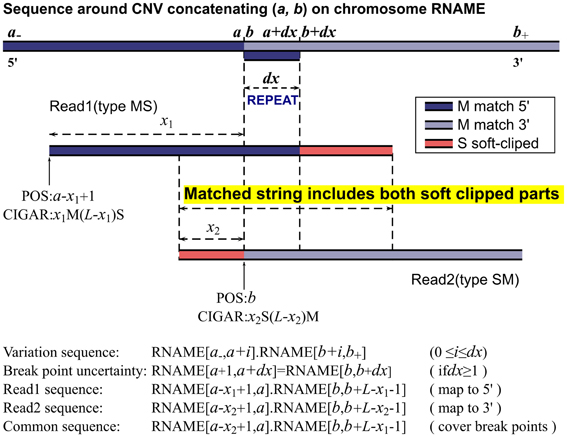
Frontiers | MATCHCLIP: locate precise breakpoints for copy number variation using CIGAR string by matching soft clipped reads