Detecting Illumina adapters by complete overlap: (a) Read 1 (R1) and... | Download Scientific Diagram
ILLUMINA READ DATA PARSING 1. Background 1.1. What is read parsing? Read parsing is the process of extracting the sequencing rea

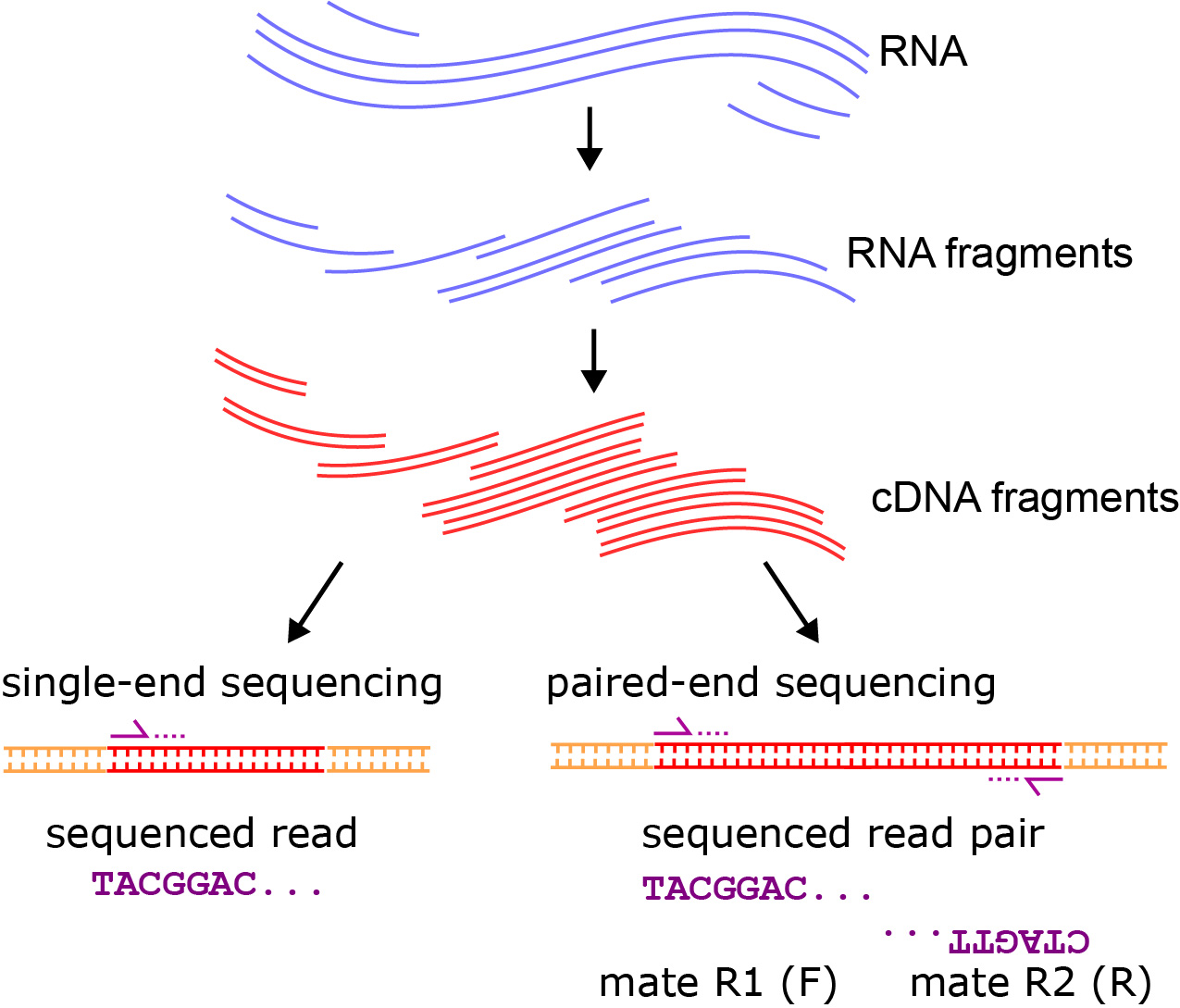
Paired-end sequencing; the inner distance between paired reads (R1 and... | Download Scientific Diagram
Quality assessment of raw FASTQ files of 100 bp paired-end reads. R1 is... | Download Scientific Diagram
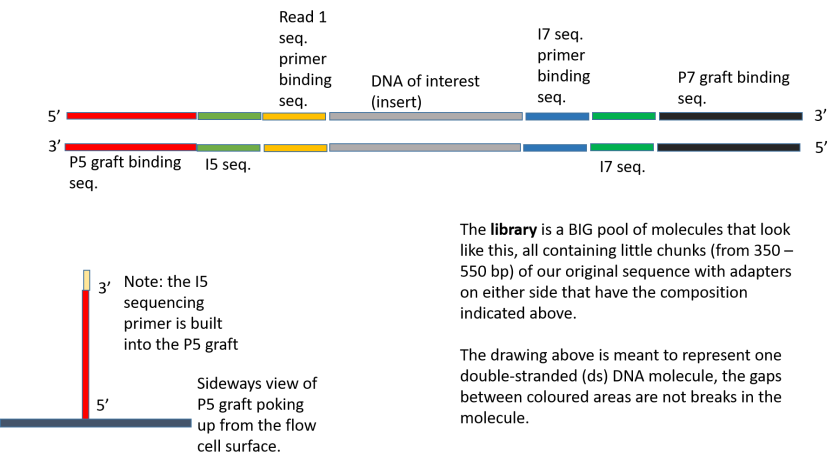
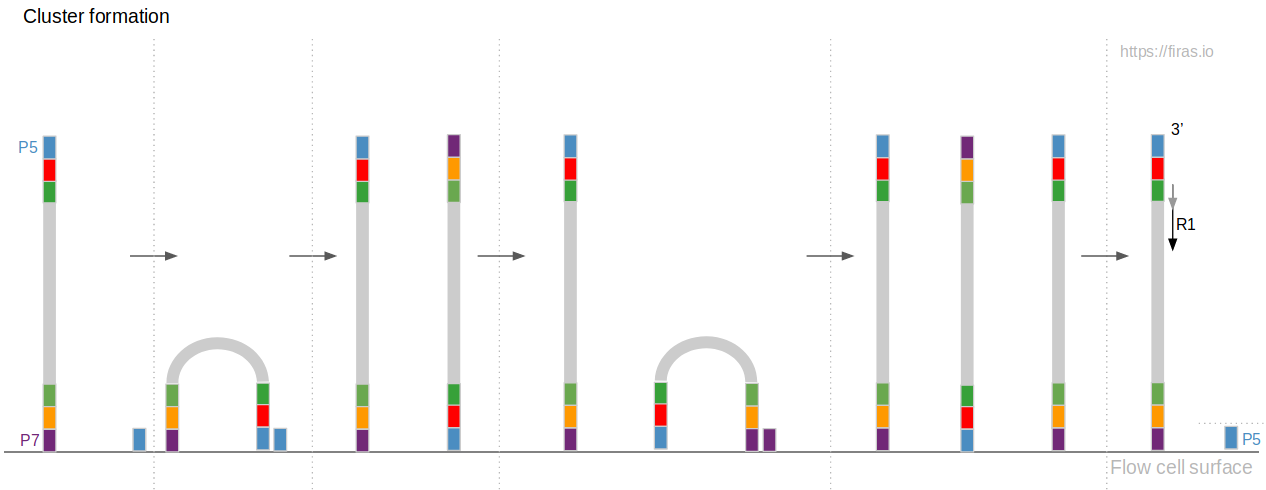
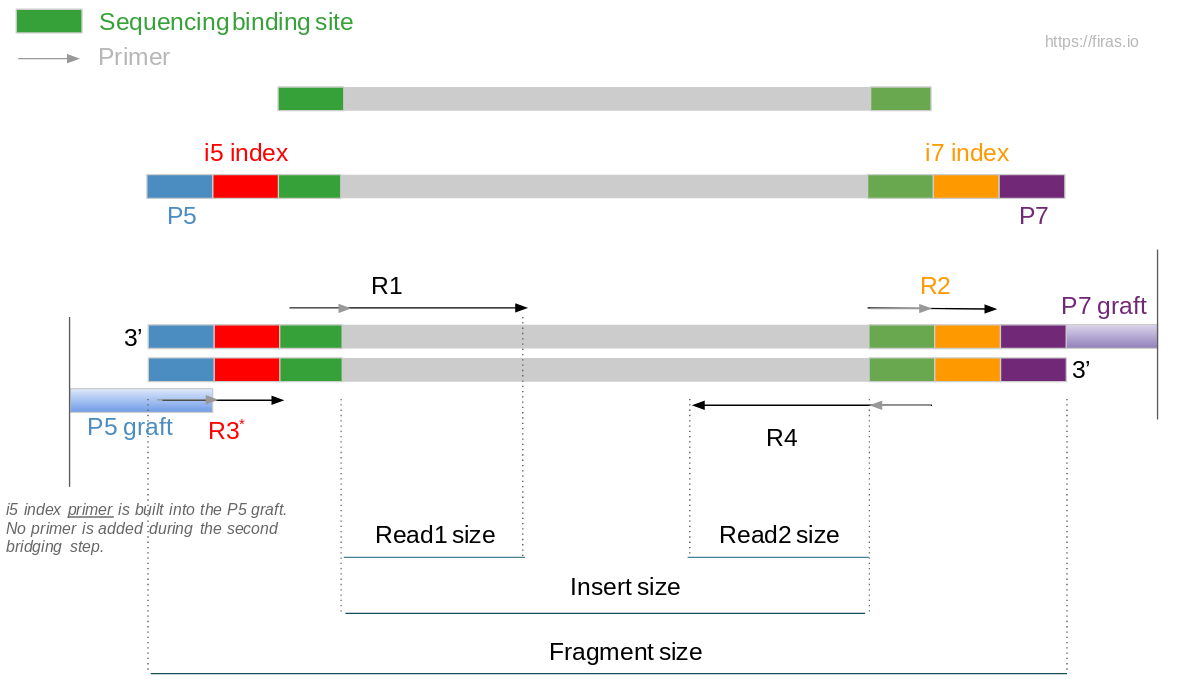
Illumina Sequencing (for Dummies) -An overview on how our samples are sequenced. – kscbioinformatics
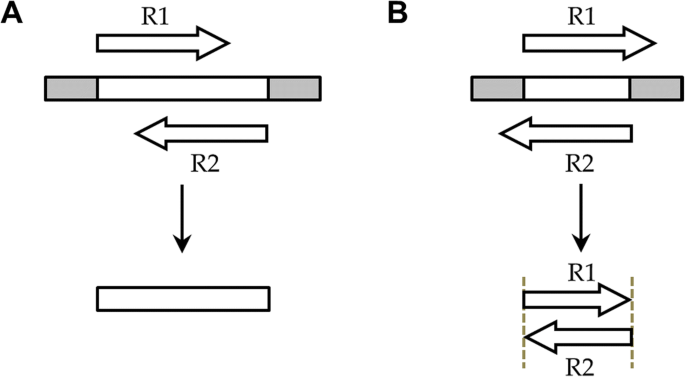
NGmerge: merging paired-end reads via novel empirically-derived models of sequencing errors | BMC Bioinformatics | Full Text
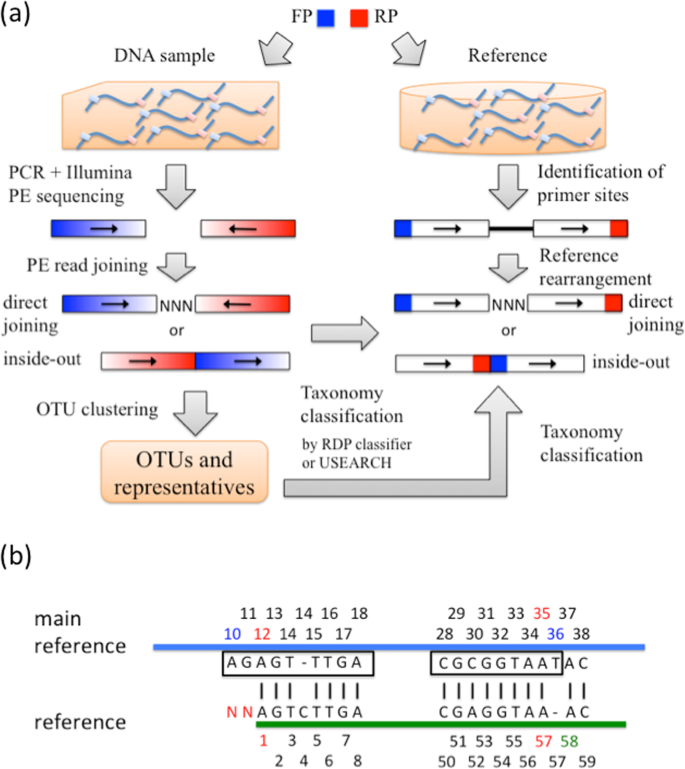
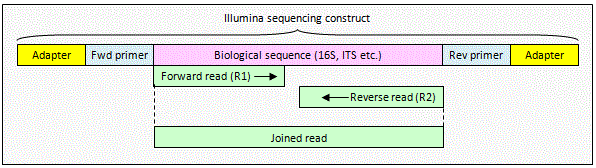
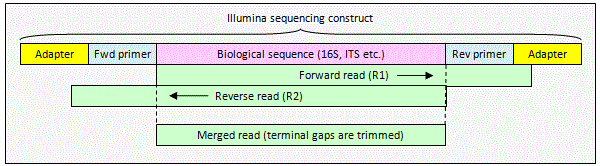
Joining Illumina paired-end reads for classifying phylogenetic marker sequences | BMC Bioinformatics | Full Text

How to quantify the overlapping reads in paired-end DNA sequencing to check the sequencing efficiency