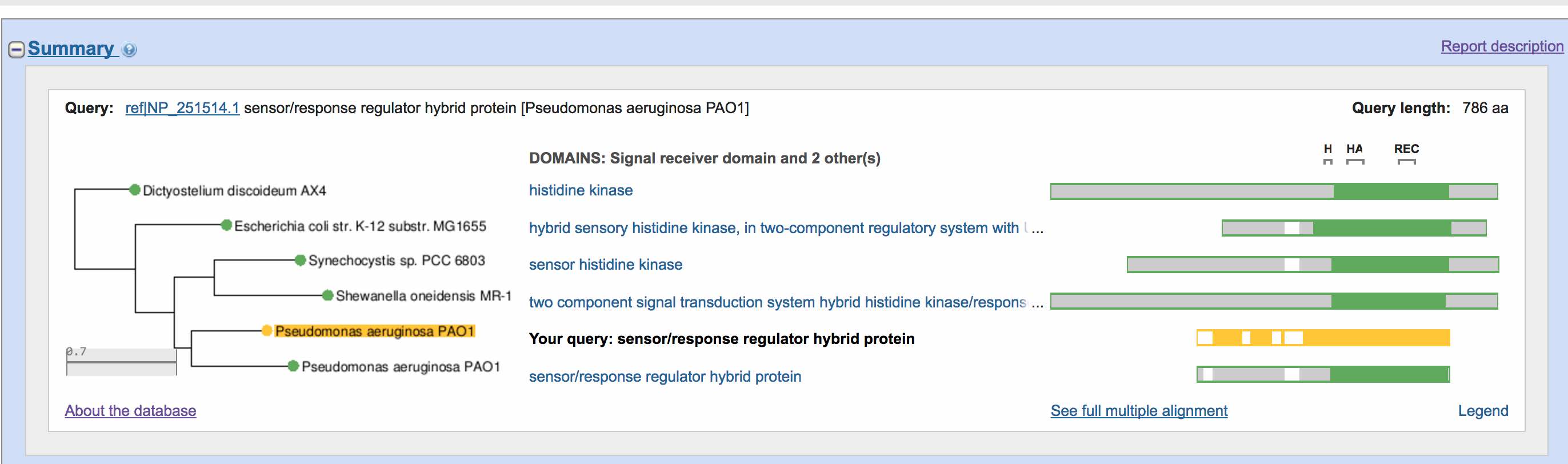
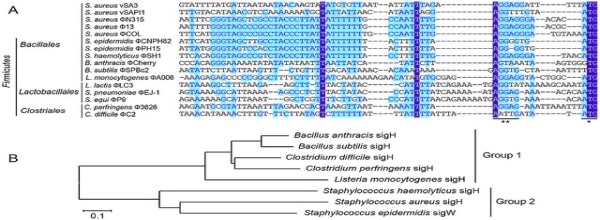
An example workflow from NCBI BLAST+ to Clustal Omega and construction... | Download Scientific Diagram

Figure 2, The expanded Dot Matrix view from Blast 2 Sequences showing the alignment of two Salmonella enterica subsp. enterica genome sequences (serovar Heidelberg str. SL476, accession NC_001083 and serovar Typhi Ty2,

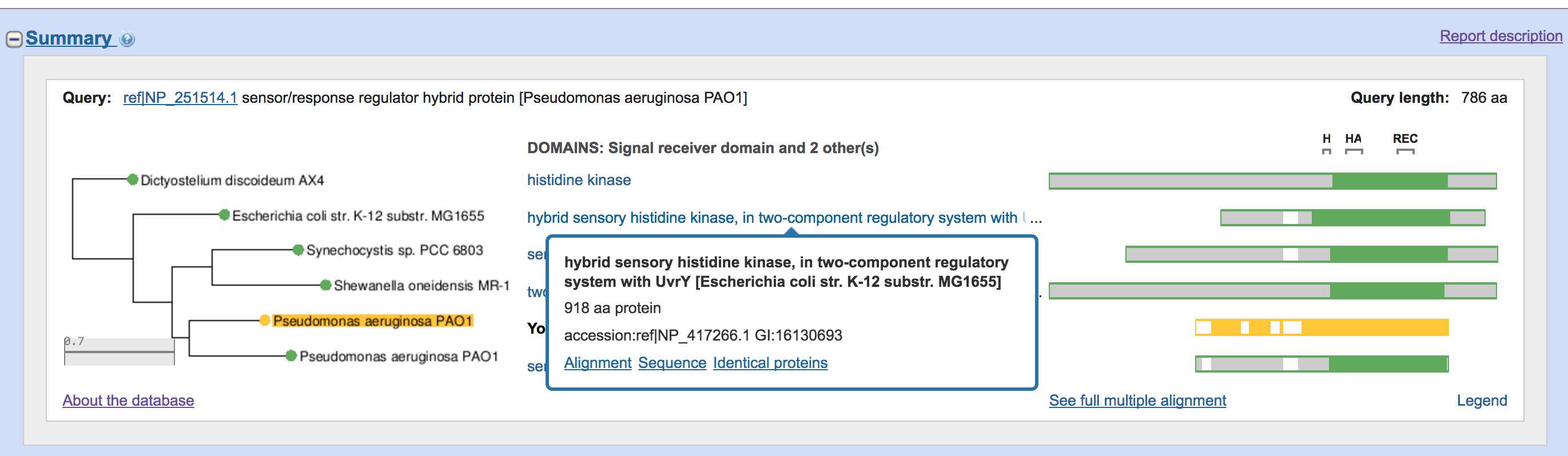
BLAST: Compare & identify sequences - NCBI Bioinformatics Resources: An Introduction - Library Guides at UC Berkeley

BLAST: Compare & identify sequences - NCBI Bioinformatics Resources: An Introduction - Library Guides at UC Berkeley