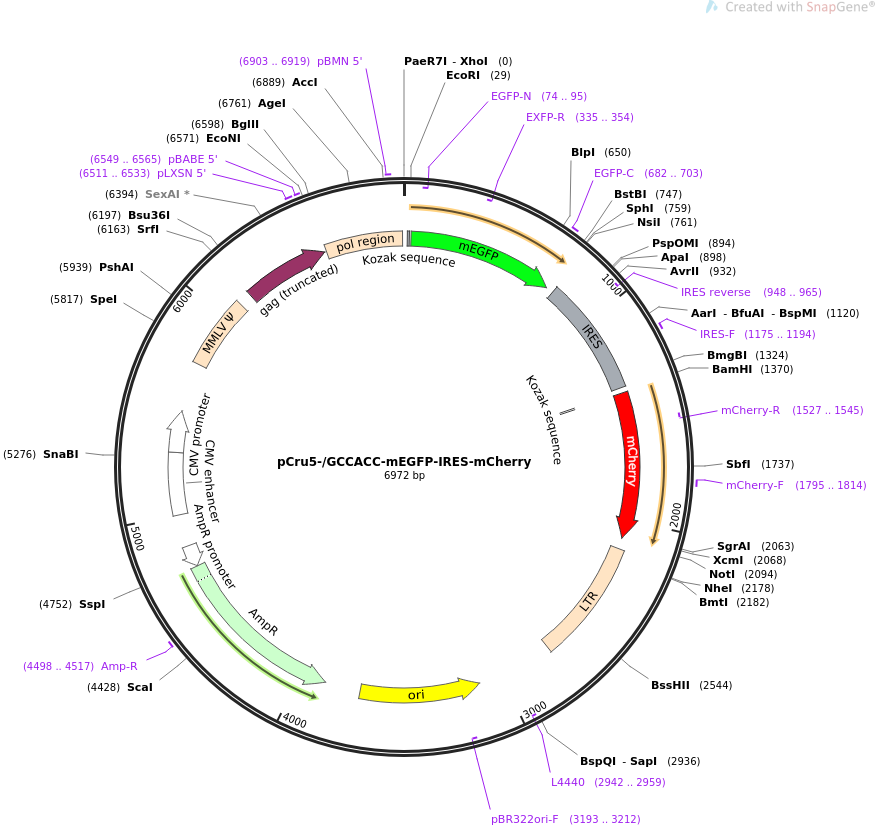
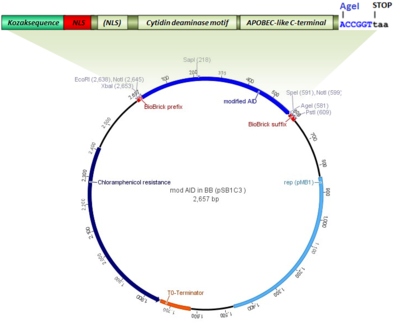
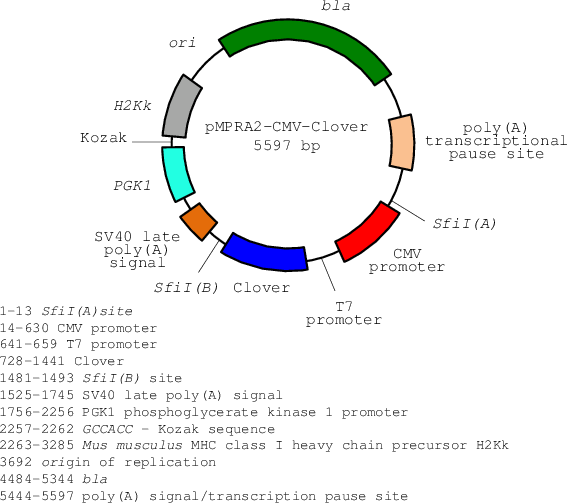
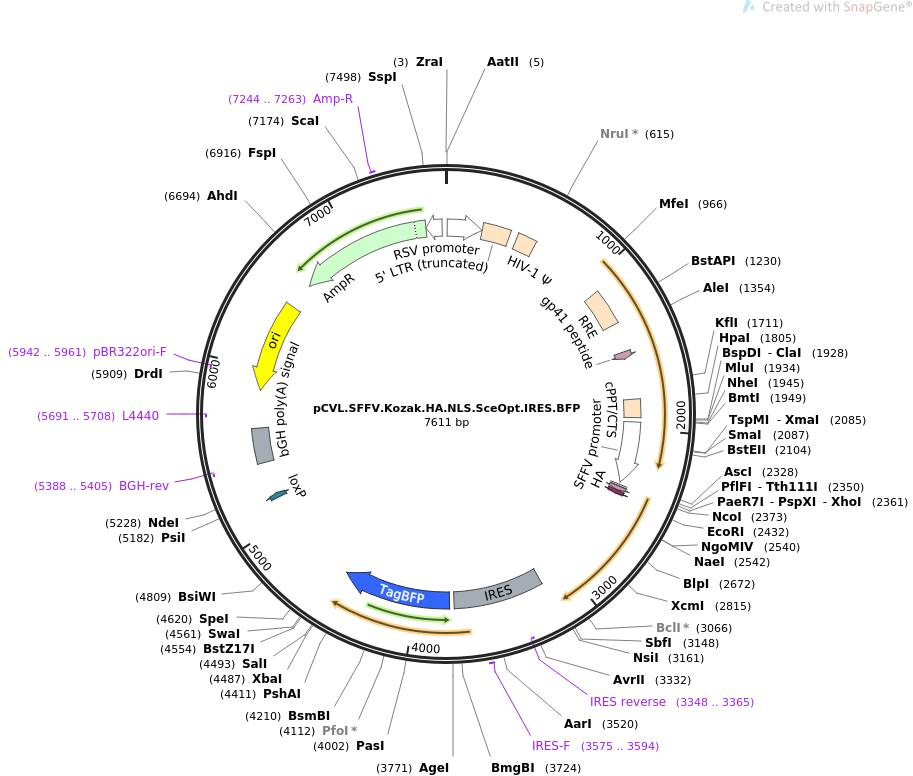
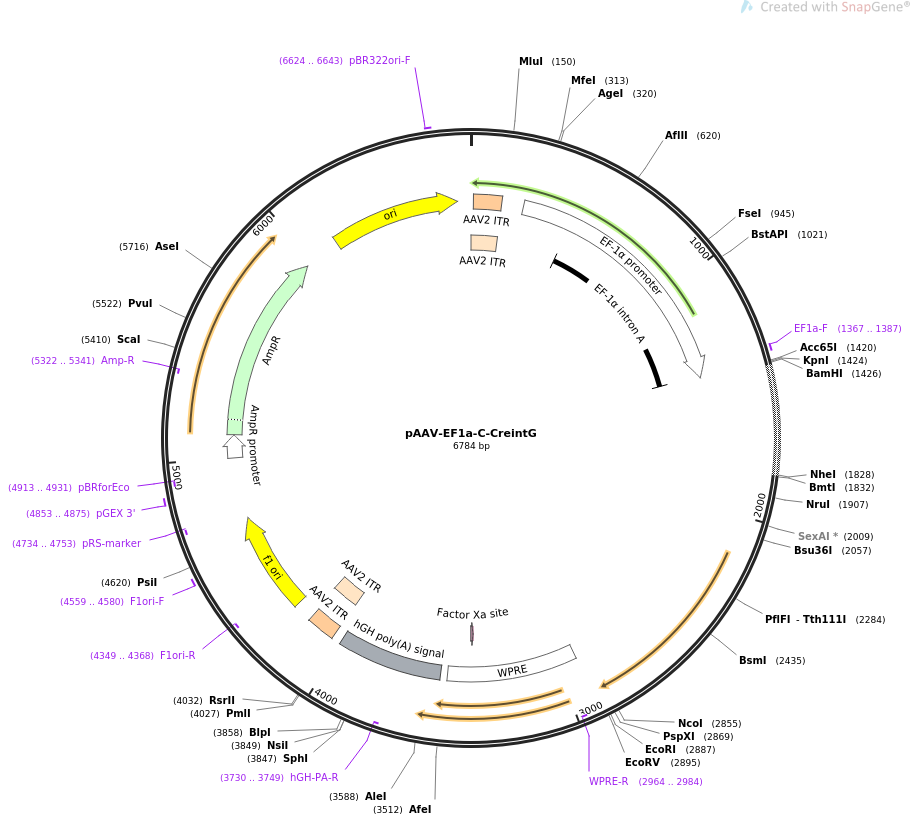
Kozak sequence improved the expression of 2A downstream gene by its... | Download Scientific Diagram
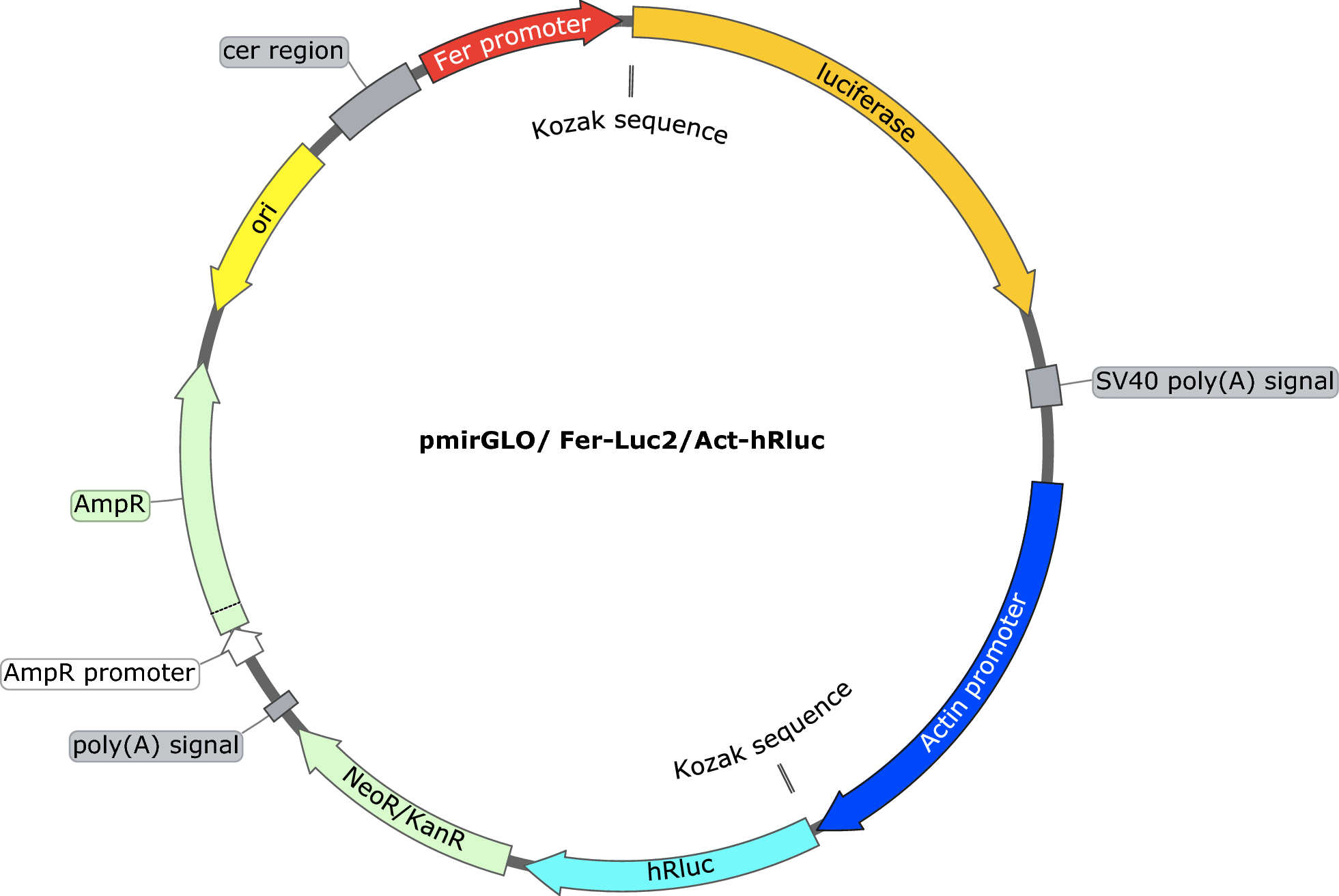
Characterization of an iron-inducible Haemaphysalis longicornis tick-derived promoter in an Ixodes scapularis-derived tick cell line (ISE6) | Parasites & Vectors | Full Text
International Genetically Engineered Machine Kozak Consensus Sequence Plasmid Nucleic Acid Sequence PNG, Clipart, African Clawed Frog,

Structural insights into the Kozak sequence interactions within the mammalian late-stage initiation complexes | bioRxiv

Binding of La protein requires a strong Kozak sequence. (A) Scheme of... | Download Scientific Diagram

A, Schema of the Kozak sequence around the initiation codon of the core... | Download Scientific Diagram
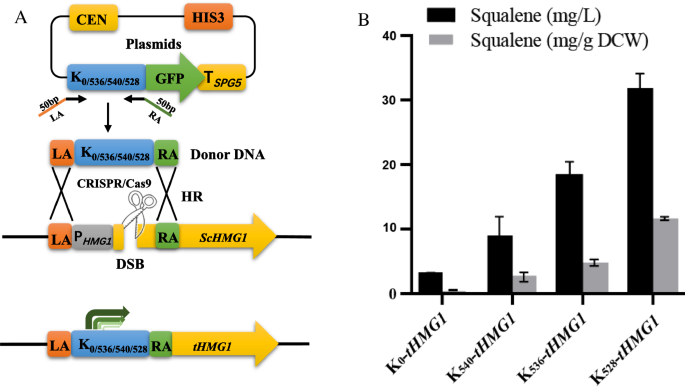
Fine-tuning the expression of pathway gene in yeast using a regulatory library formed by fusing a synthetic minimal promoter with different Kozak variants | Microbial Cell Factories | Full Text

What is the Difference Between Shine Dalgarno and Kozak Sequence | Compare the Difference Between Similar Terms

International Genetically Engineered Machine Kozak Consensus Sequence Plasmid Nucleic Acid Sequence, PNG, 2366x917px, Kozak Consensus Sequence,