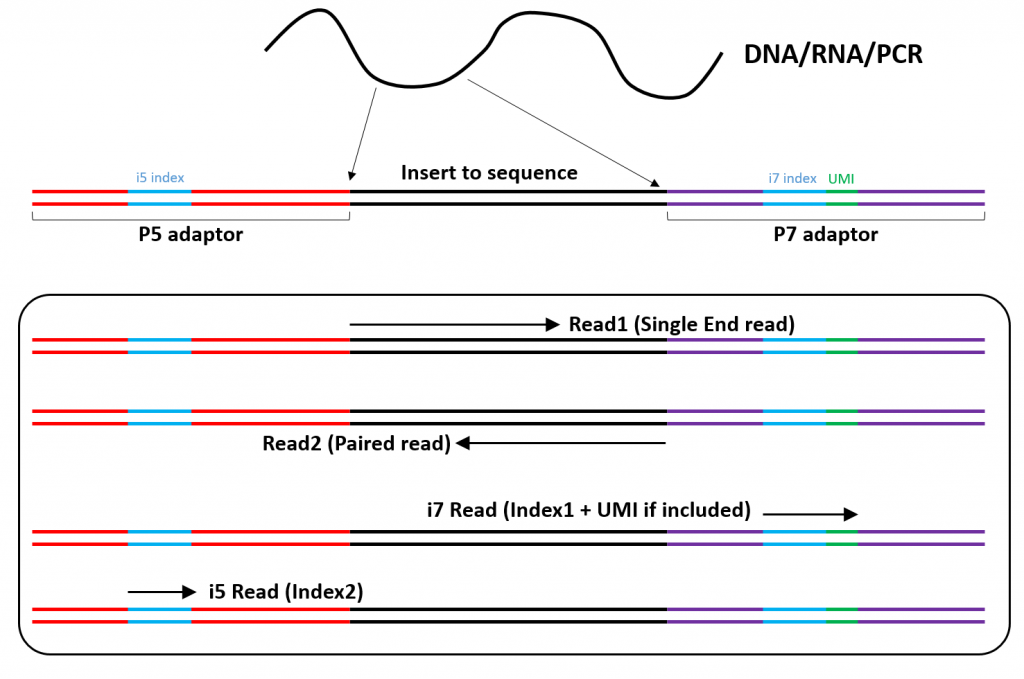
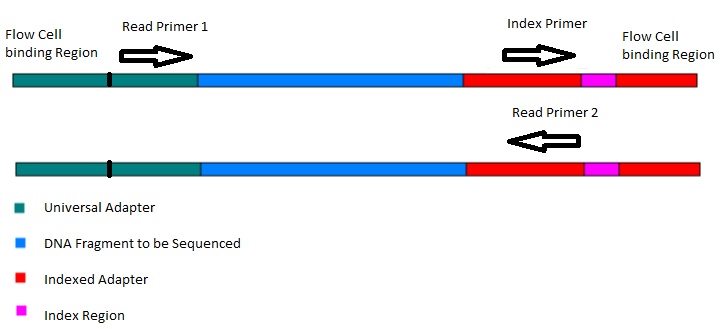
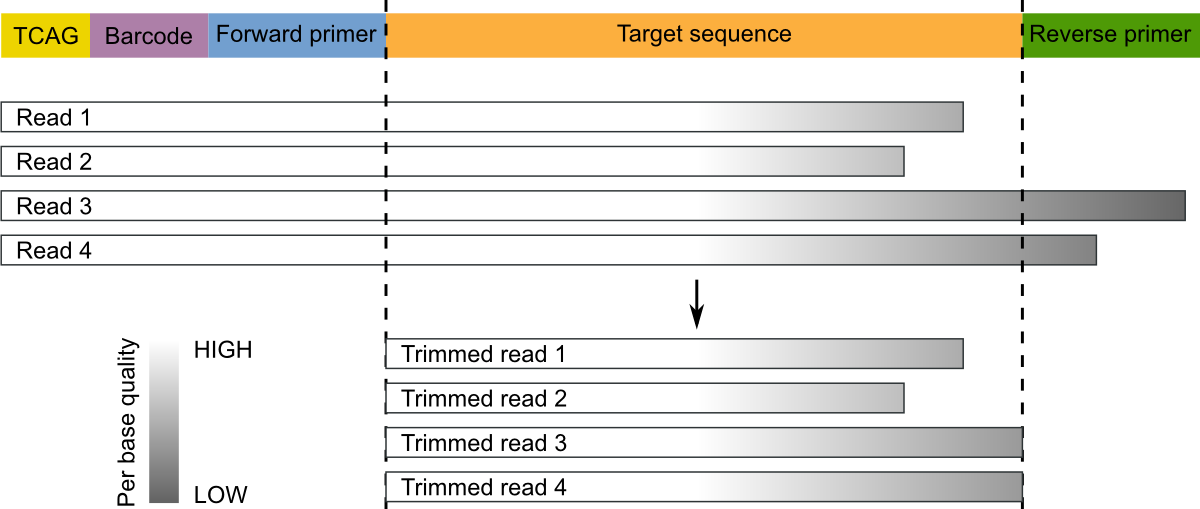
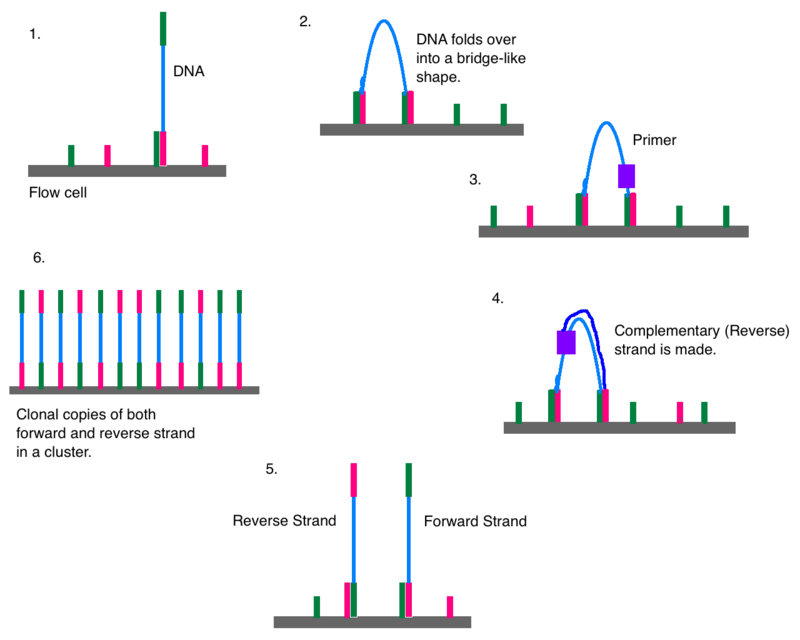
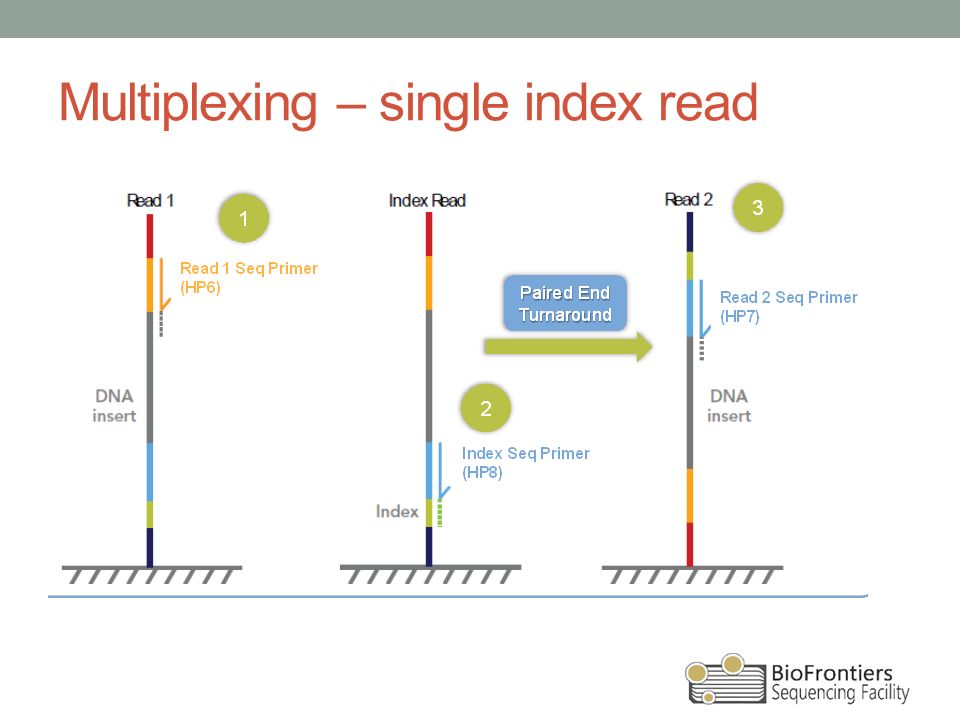
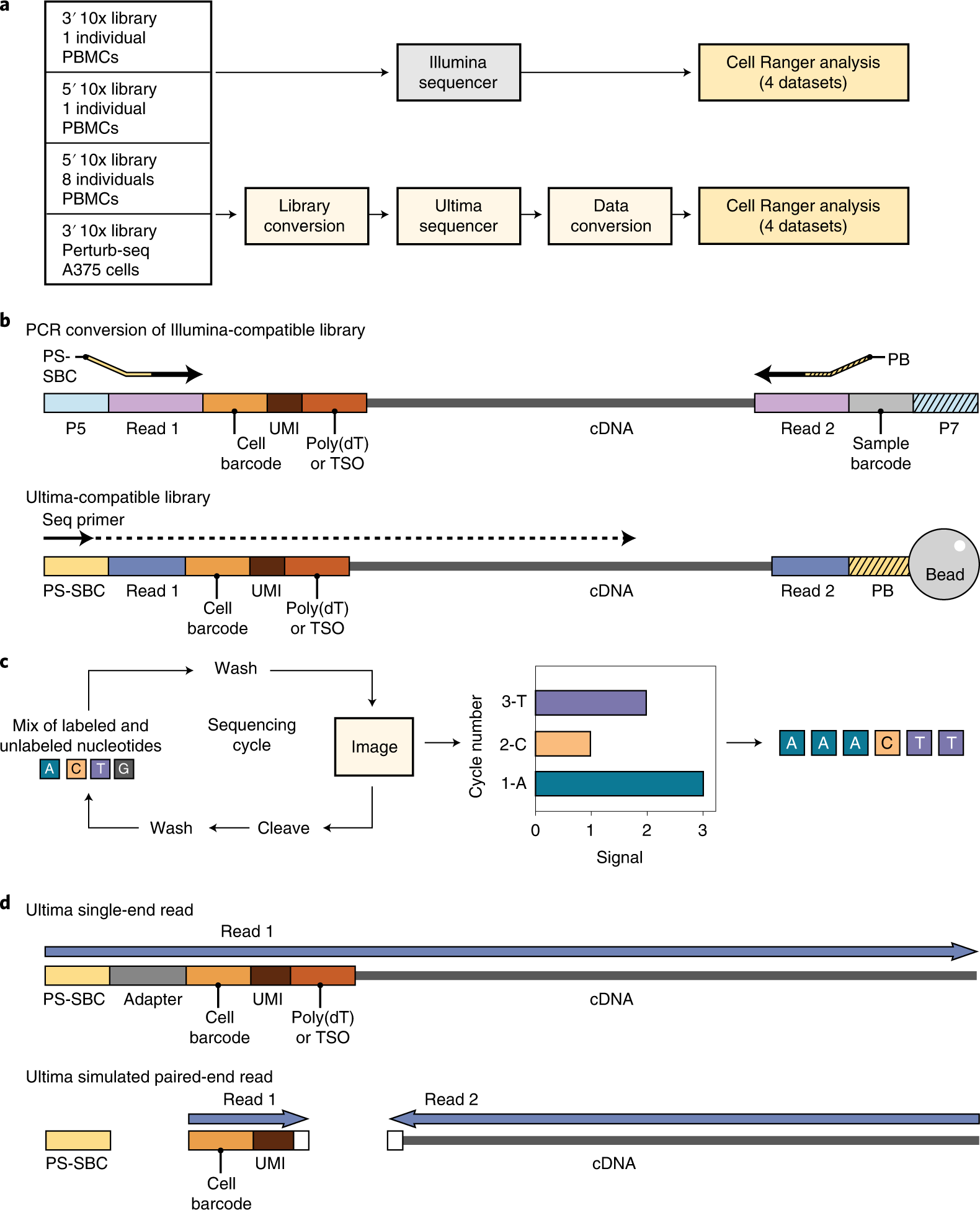
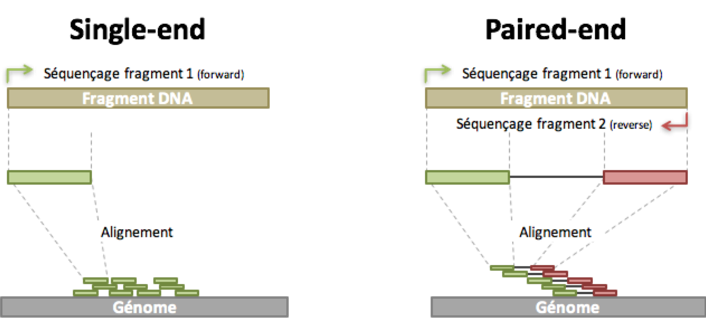
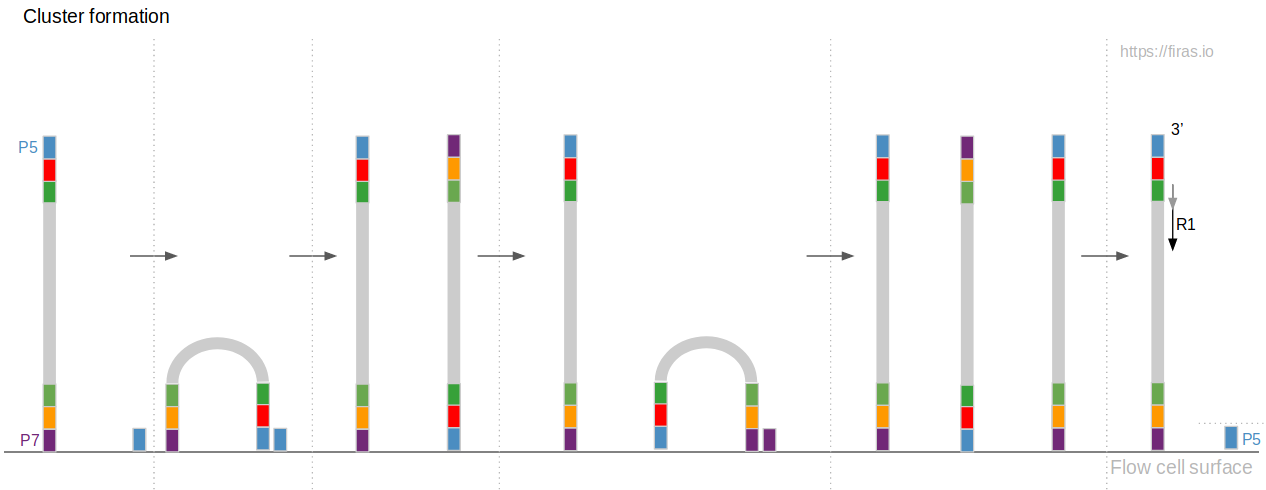
Paired-end sequencing (left) showing Read 1 and Read 2 primers starting... | Download Scientific Diagram
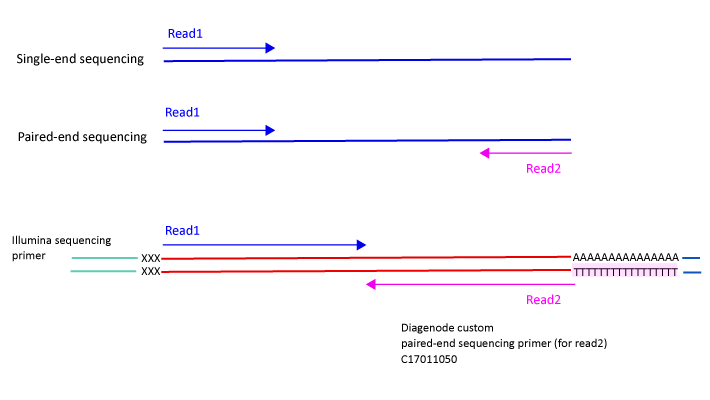
Library preparation for RNA sequencing utilizing - Capture and Amplification by Tailing and Switching (CATS) | Diagenode
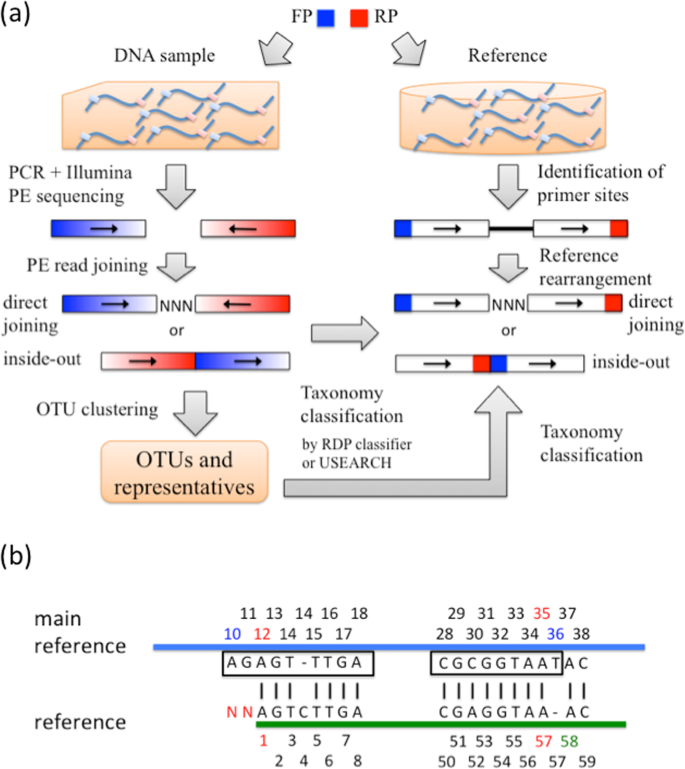
Joining Illumina paired-end reads for classifying phylogenetic marker sequences | BMC Bioinformatics | Full Text