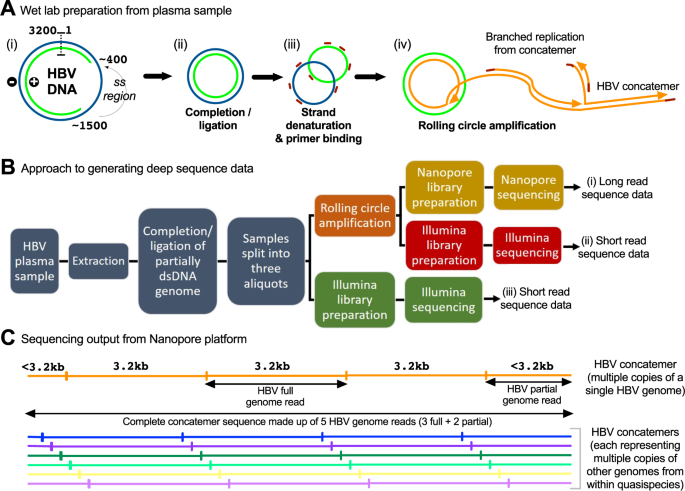
Illumina and Nanopore methods for whole genome sequencing of hepatitis B virus (HBV) | Scientific Reports

Histogram of read variants' lengths in Illumina sequencing data. The... | Download Scientific Diagram

The distribution of read lengths in 454, Illumina and PacBio budgerigar... | Download Scientific Diagram
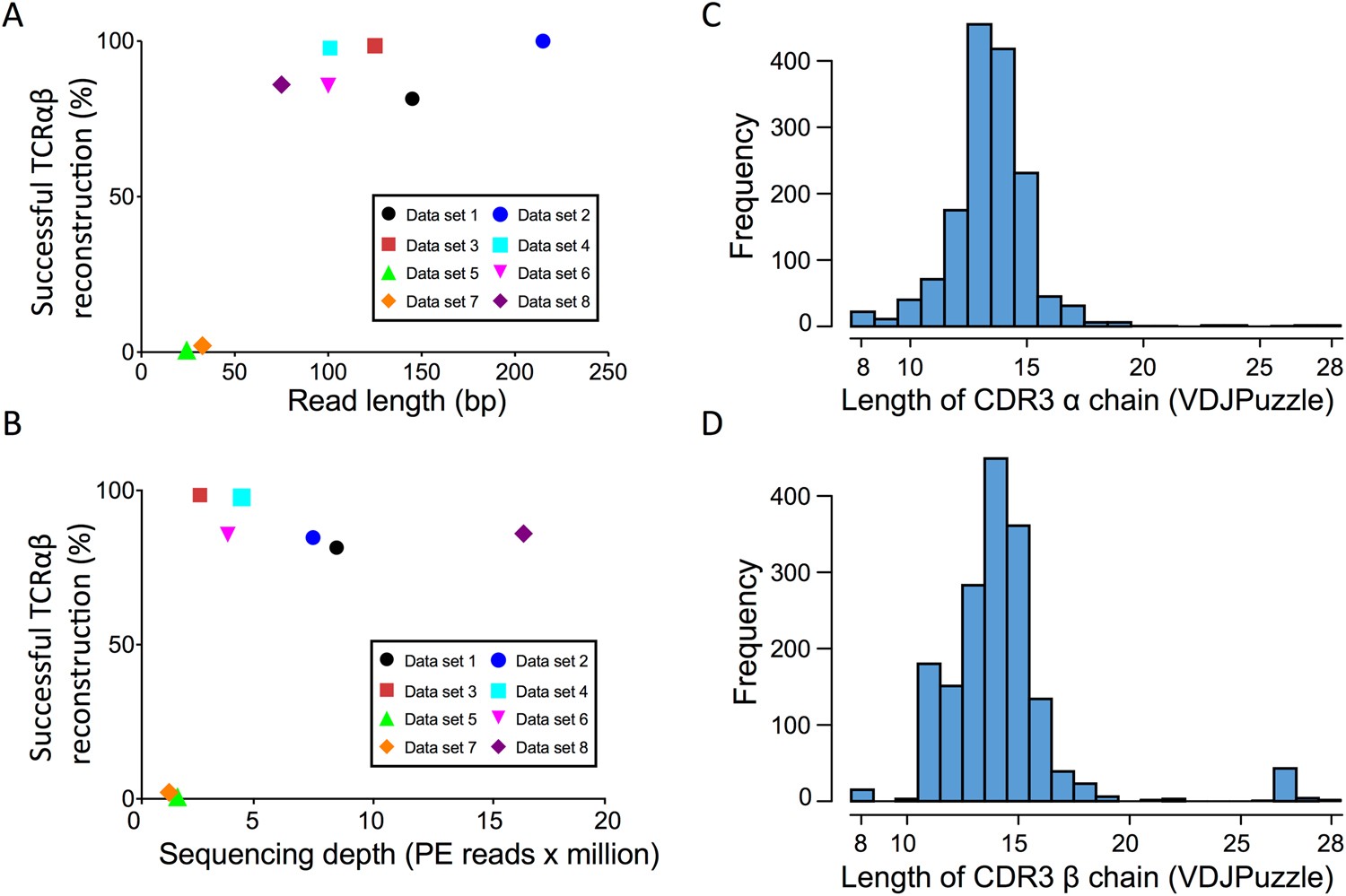
Impact of sequencing depth and read length on single cell RNA sequencing data of T cells | Scientific Reports

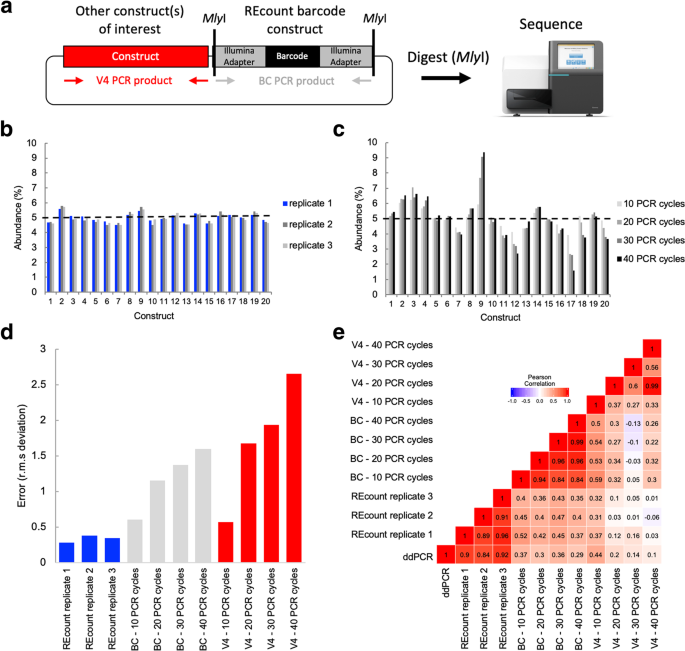
Ultrahigh-Throughput Multiplexing and Sequencing of >500-Base-Pair Amplicon Regions on the Illumina HiSeq 2500 Platform | mSystems
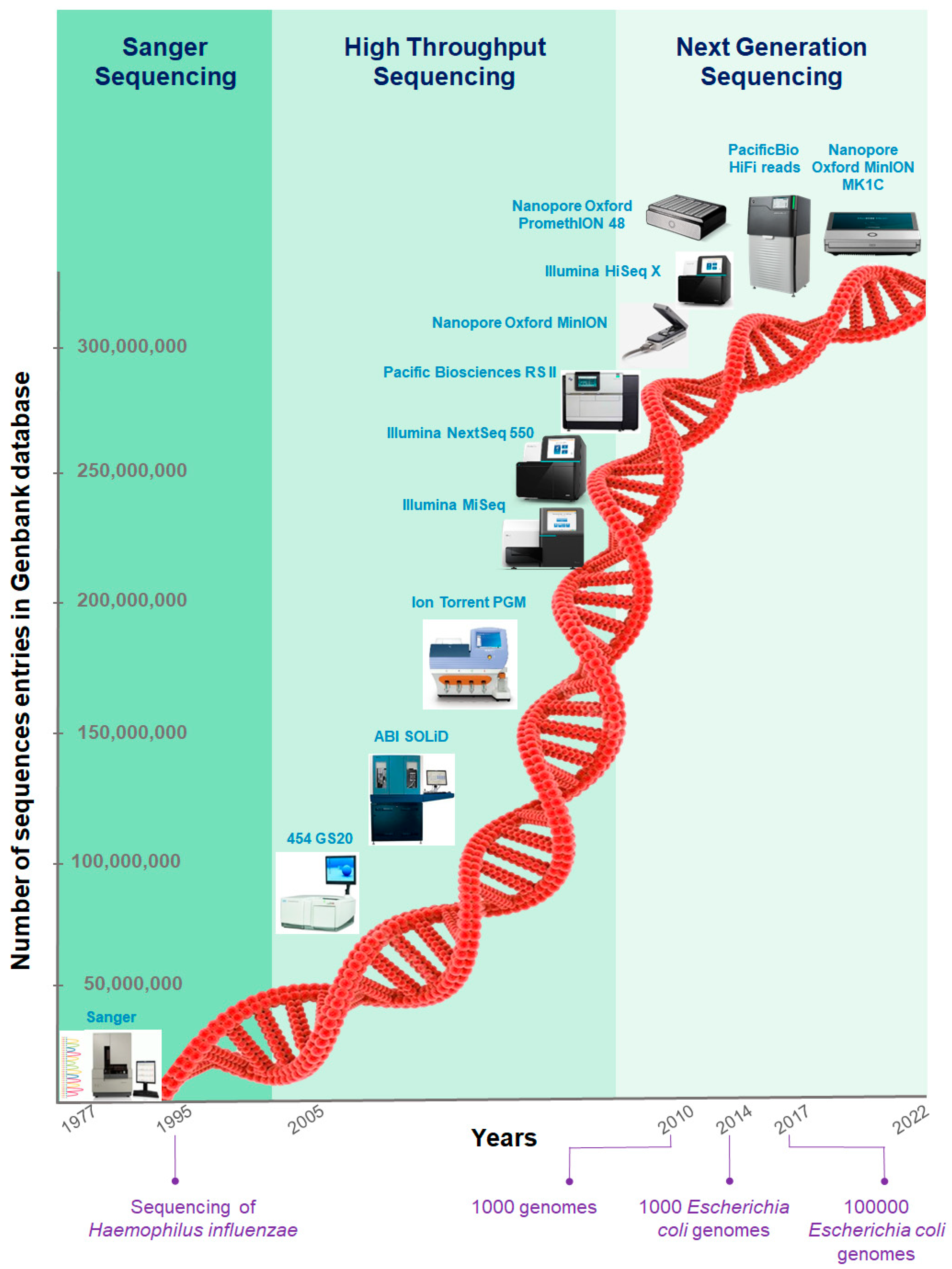
IJMS | Free Full-Text | Application and Challenge of 3rd Generation Sequencing for Clinical Bacterial Studies
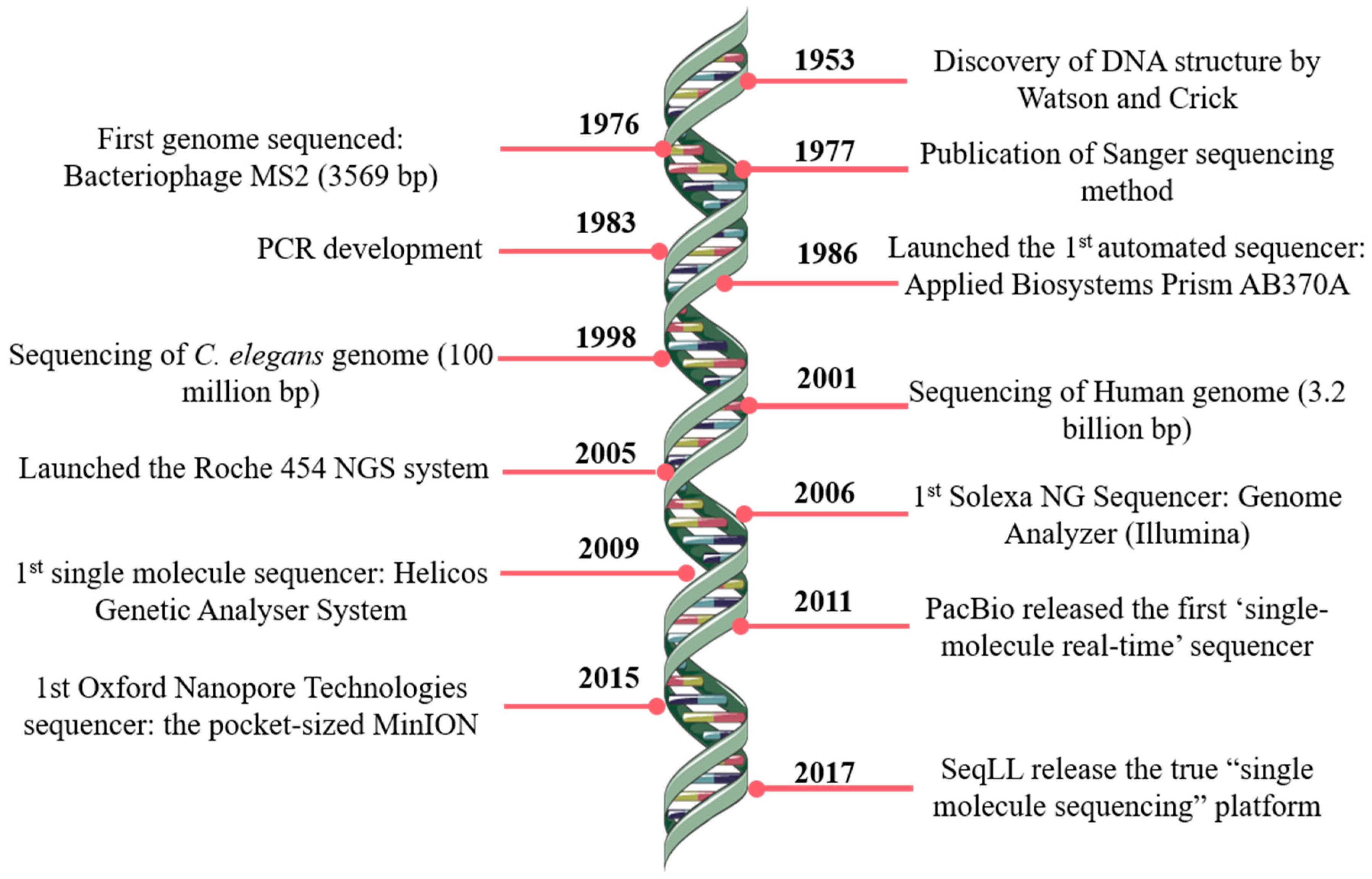
JCM | Free Full-Text | Bioinformatics and Computational Tools for Next-Generation Sequencing Analysis in Clinical Genetics