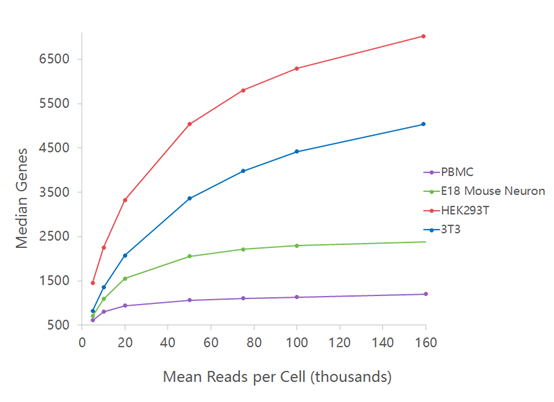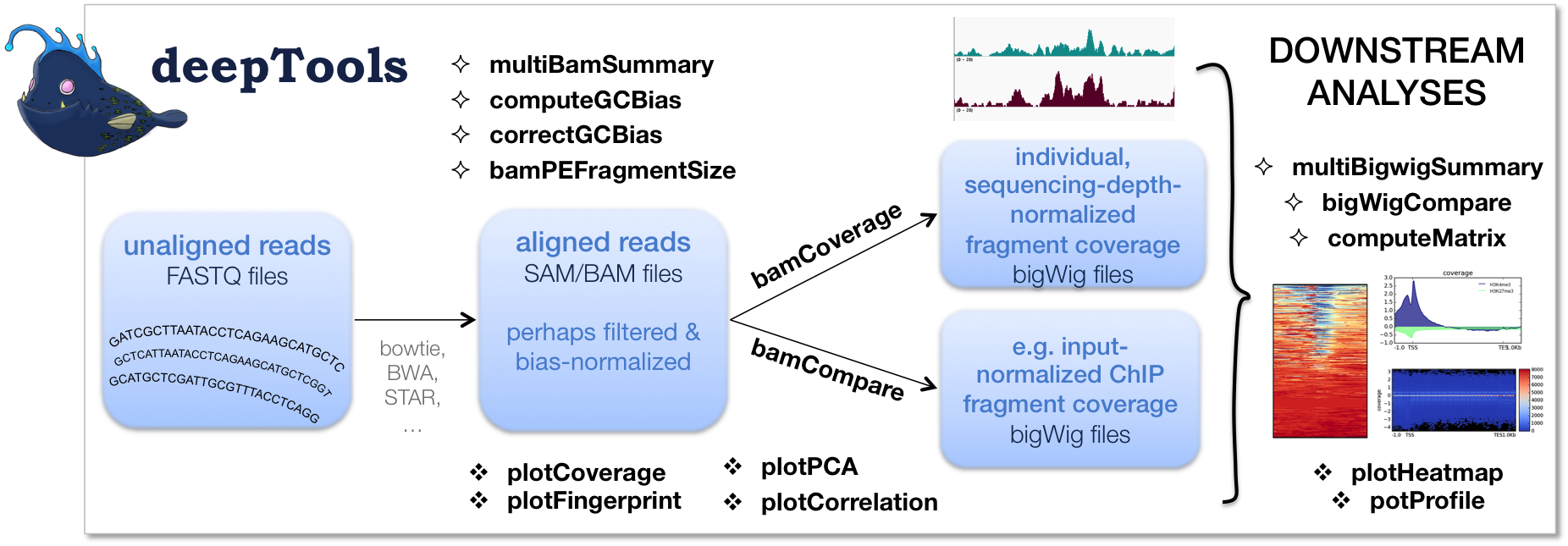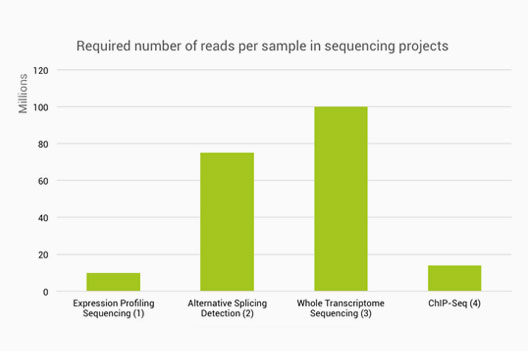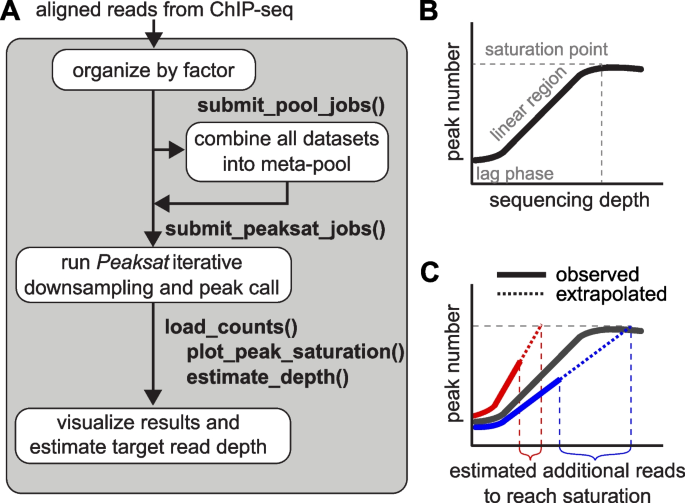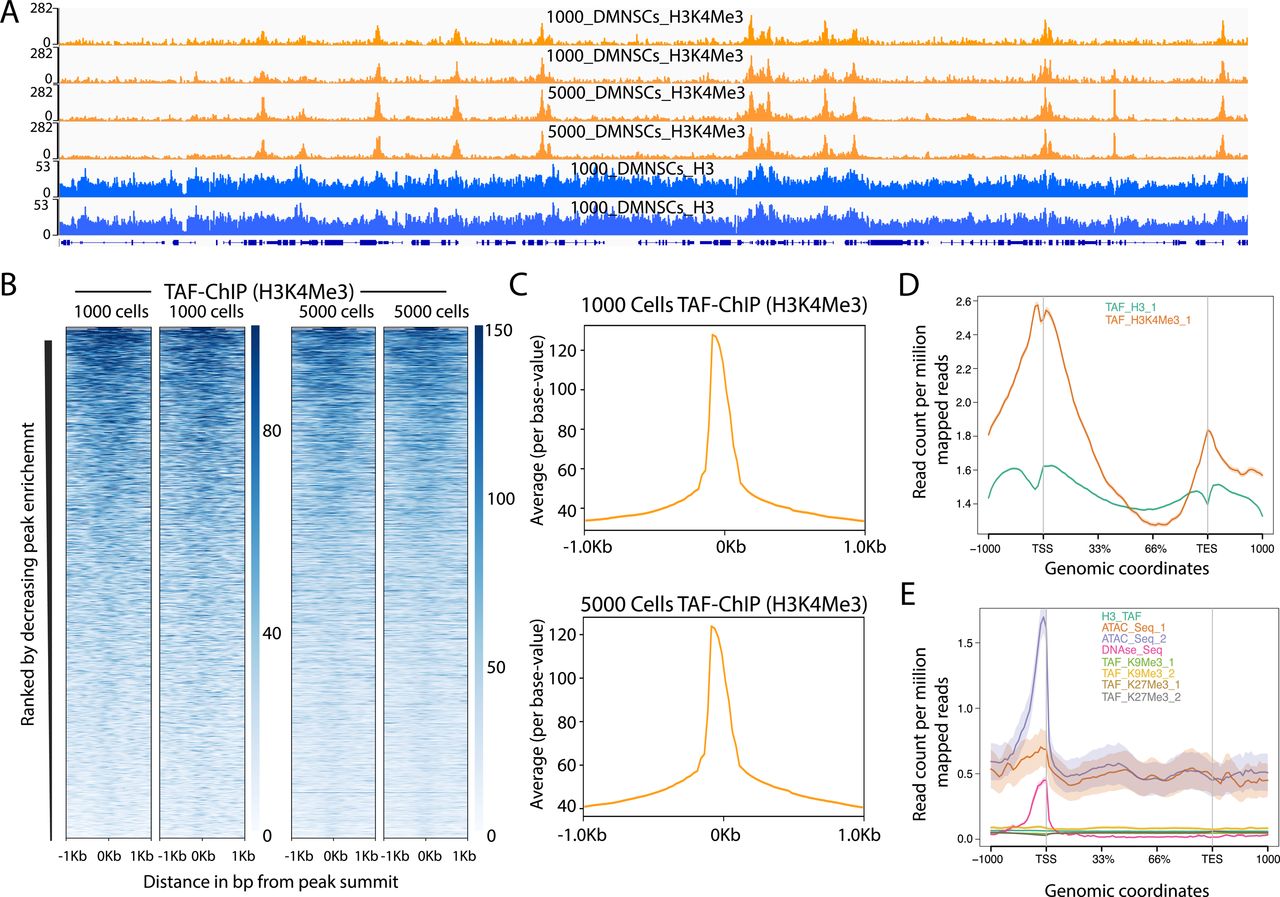
TAF-ChIP: an ultra-low input approach for genome-wide chromatin immunoprecipitation assay | Life Science Alliance

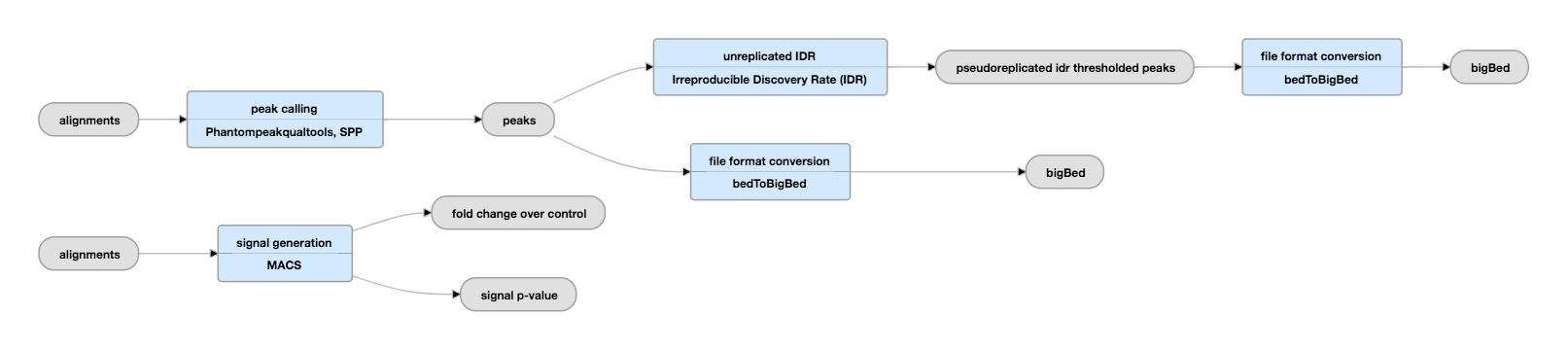
Podstawy komputerowej analizy danych pochodzących z ChIP-seq :: Biotechnologia: e-biotechnologia.pl :: Biotechnologiczny Portal Internetowy-aktualności, artykuły, laboratorium, studia biotechnologiczne.